Protein c-Fos
Protein c-Fos is a proto-oncogene that is the human homolog of the retroviral oncogene v-fos.[5] It is encoded in humans by the FOS gene. It was first discovered in rat fibroblasts as the transforming gene of the FBJ MSV (Finkel–Biskis–Jinkins murine osteogenic sarcoma virus) (Curran and Tech, 1982). It is a part of a bigger Fos family of transcription factors which includes c-Fos, FosB, Fra-1 and Fra-2.[6] It has been mapped to chromosome region 14q21→q31. c-Fos encodes a 62 kDa protein, which forms heterodimer with c-jun (part of Jun family of transcription factors), resulting in the formation of AP-1 (Activator Protein-1) complex which binds DNA at AP-1 specific sites at the promoter and enhancer regions of target genes and converts extracellular signals into changes of gene expression.[7] It plays an important role in many cellular functions and has been found to be overexpressed in a variety of cancers.
Structure and function
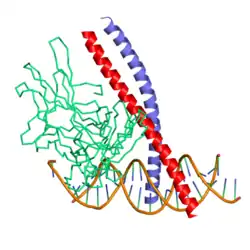
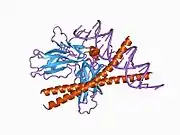
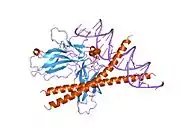
c-Fos is a 380 amino acid protein with a basic leucine zipper region for dimerisation and DNA-binding and a transactivation domain at C-terminus, and, like Jun proteins, it can form homodimers.[8] In vitro studies have shown that Jun–Fos heterodimers are more stable and have stronger DNA-binding activity than Jun–Jun homodimers.[9]
A variety of stimuli, including serum, growth factors, tumor promoters, cytokines, and UV radiation induce their expression. The c-fos mRNA and protein is generally among the first to be expressed and hence referred to as an immediate early gene. It is rapidly and transiently induced, within 15 minutes of stimulation.[10] Its activity is also regulated by posttranslational modification caused by phosphorylation by different kinases, like MAPK, CDC2, PKA or PKC which influence protein stability, DNA-binding activity and the trans-activating potential of the transcription factors.[11][12][13] It can cause gene repression as well as gene activation, although different domains are believed to be involved in both processes.
It is involved in important cellular events, including cell proliferation, differentiation and survival; genes associated with hypoxia; and angiogenesis;[14] which makes its dysregulation an important factor for cancer development. It can also induce a loss of cell polarity and epithelial-mesenchymal transition, leading to invasive and metastatic growth in mammary epithelial cells.[15]
The importance of c-fos in biological context has been determined by eliminating endogenous function by using anti-sense mRNA, anti-c-fos antibodies, a ribozyme that cleaves c-fos mRNA or a dominant negative mutant of c-fos. The transgenic mice thus generated are viable, demonstrating that there are c-fos dependent and independent pathways of cell proliferation, but display a range of tissue-specific developmental defects, including osteoporosis, delayed gametogenesis, lymphopenia and behavioral abnormalities.
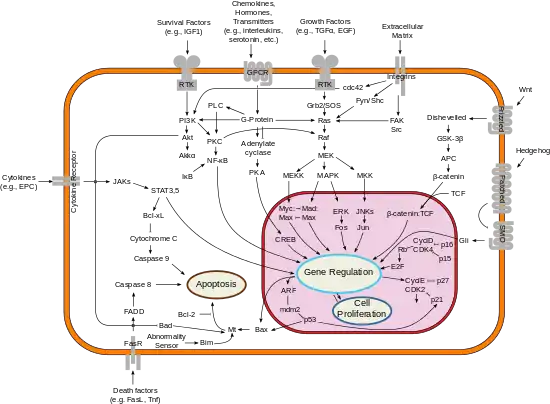
Clinical significance
Signaling cascade in the nucleus accumbens that results in psychostimulant addiction
|
The AP-1 complex has been implicated in transformation and progression of cancer. In osteosarcoma and endometrial carcinoma, c-Fos overexpression was associated with high-grade lesions and poor prognosis. Also, in a comparison between precancerous lesion of the cervix uteri and invasive cervical cancer, c-Fos expression was significantly lower in precancerous lesions. c-Fos has also been identified as independent predictor of decreased survival in breast cancer.[23]
It was found that overexpression of c-fos from class I MHC promoter in transgenic mice leads to the formation of osteosarcomas due to increased proliferation of osteoblasts whereas ectopic expression of the other Jun and Fos proteins does not induce any malignant tumors. Activation of the c-Fos transgene in mice results in overexpression of cyclin D1, A and E in osteoblasts and chondrocytes, both in vitro and in vivo, which might contribute to the uncontrolled growth leading to tumor. Human osteosarcomas analyzed for c-fos expression have given positive results in more than half the cases and c-fos expression has been associated with higher frequency of relapse and poor response to chemotherapy.
Several studies have raised the idea that c-Fos may also have tumor-suppressor activity, that it might be able to promote as well as suppress tumorigenesis. Supporting this is the observation that in ovarian carcinomas, loss of c-Fos expression correlates with disease progression. This double action could be enabled by differential protein composition of tumour cells and their environment, for example, dimerisation partners, co-activators and promoter architecture. It is possible that the tumor suppressing activity is due to a proapoptotic function. The exact mechanism by which c-Fos contributes to apoptosis is not clearly understood, but observations in human hepatocellular carcinoma cells indicate that c-Fos is a mediator of c-myc-induced cell death and might induce apoptosis through the p38 MAP kinase pathway. Fas ligand (FASLG or FasL) and the tumour necrosis factor-related apoptosis-inducing ligand (TNFSF10 or TRAIL) might reflect an additional apoptotic mechanism induced by c-Fos, as observed in a human T-cell leukaemia cell line. Another possible mechanism of c-Fos involvement in tumour suppression could be the direct regulation of BRCA1, a well established factor in familial breast and ovarian cancer.
In addition, the role of c-fos and other Fos family proteins has also been studied in endometrial carcinoma, cervical cancer, mesotheliomas, colorectal cancer, lung cancer, melanomas, thyroid carcinomas, esophageal cancer, hepatocellular carcinomas, etc.
Cocaine, methamphetamine,[24] morphine,[25] and other psychoactive drugs[26][27] have been shown to increase c-Fos production in the mesocortical pathway (prefrontal cortex) as well as in the mesolimbic reward pathway (nucleus accumbens), as well as display variability depending on prior sensitization.[27] c-Fos repression by ΔFosB's AP-1 complex within the D1-type medium spiny neurons of the nucleus accumbens acts as a molecular switch that enables the chronic induction of ΔFosB, thus allowing it to accumulate more rapidly. As such, the c-Fos promoter finds utilization in drug addiction research in general, as well as with context-induced relapse to drug-seeking and other behavioral changes associated with chronic drug taking.
An increase in c-Fos production in androgen receptor-containing neurons has been observed in rats after mating.
Applications
Expression of c-fos is an indirect marker of neuronal activity because c-fos is often expressed when neurons fire action potentials.[28][29] Upregulation of c-fos mRNA in a neuron indicates recent activity.[30]
The c-fos promoter has also been utilised for drug abuse research. Scientists use this promoter to turn on transgenes in rats, allowing them to manipulate specific neuronal ensembles to assess their role in drug-related memories and behavior.[31] This neuronal control can be replicated with optogenetics or DREADDs [32]
Interactions
c-Fos has been shown to interact with:
- BCL3,[33]
- COBRA1,[34]
- CSNK2A1,[35]
- CSNK2A2,[35]
- DDIT3,[36]
- JUN[37][38][39][40][41][42][43]
- NCOA1;,[44][45]
- NCOR2,[46]
- RELA,[37]
- RUNX1,[47][48]
- RUNX2,[47][48]
- SMAD3,[49] and
- TBP.[50]
 |

See also
- Leptomycin
- c-Jun
- Egr-1
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000170345 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000021250 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ Curran, T: The c-fos proto-oncogene. In: Reddy EP, Skalka AM, Curran T (eds.). The Oncogene Handbook 1988 Elsevier, New York, pp 307–327
- ↑ Milde-Langosch K (November 2005). "The Fos family of transcription factors and their role in tumourigenesis". Eur. J. Cancer. 41 (16): 2449–61. doi:10.1016/j.ejca.2005.08.008. PMID 16199154.
- ↑ Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M (August 1988). "The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes". Cell. 54 (4): 541–52. doi:10.1016/0092-8674(88)90076-1. PMID 3135940. S2CID 43078284.
- ↑ Szalóki N, Krieger JW, Komáromi I, Tóth K, Vámosi G (November 2015). "Evidence for homodimerization of the c-Fos transcription factor in live cells revealed by fluorescence microscopy and computer modeling". Mol. Cell. Biol. 35 (21): 3785–98. doi:10.1128/MCB.00346-15. PMC 4589601. PMID 26303532.
- ↑ Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P (December 1988). "c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities" (PDF). Cell. 55 (5): 917–24. doi:10.1016/0092-8674(88)90147-X. PMID 3142692. S2CID 19876513.
- ↑ Hu E, Mueller E, Oliviero S, Papaioannou VE, Johnson R, Spiegelman BM (July 1994). "Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes". EMBO J. 13 (13): 3094–103. doi:10.1002/j.1460-2075.1994.tb06608.x. PMC 395200. PMID 8039503.
- ↑ Gruda MC, Kovary K, Metz R, Bravo R (September 1994). "Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity". Oncogene. 9 (9): 2537–47. PMID 8058317.
- ↑ Hurd TW, Culbert AA, Webster KJ, Tavaré JM (December 2002). "Dual role for mitogen-activated protein kinase (Erk) in insulin-dependent regulation of Fra-1 (fos-related antigen-1) transcription and phosphorylation". Biochem. J. 368 (Pt 2): 573–80. doi:10.1042/BJ20020579. PMC 1223008. PMID 12197835.
- ↑ Rosenberger SF, Finch JS, Gupta A, Bowden GT (January 1999). "Extracellular signal-regulated kinase 1/2-mediated phosphorylation of JunD and FosB is required for okadaic acid-induced activator protein 1 activation". J. Biol. Chem. 274 (2): 1124–30. doi:10.1074/jbc.274.2.1124. PMID 9873060.
- ↑ Tulchinsky E (July 2000). "Fos family members: regulation, structure and role in oncogenic transformation". Histol. Histopathol. 15 (3): 921–8. PMID 10963134.
- ↑ Fialka I, Schwarz H, Reichmann E, Oft M, Busslinger M, Beug H (March 1996). "The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions". J. Cell Biol. 132 (6): 1115–32. doi:10.1083/jcb.132.6.1115. PMC 2120757. PMID 8601589.
- 1 2 3 Renthal W, Nestler EJ (September 2009). "Chromatin regulation in drug addiction and depression". Dialogues in Clinical Neuroscience. 11 (3): 257–268. PMC 2834246. PMID 19877494.
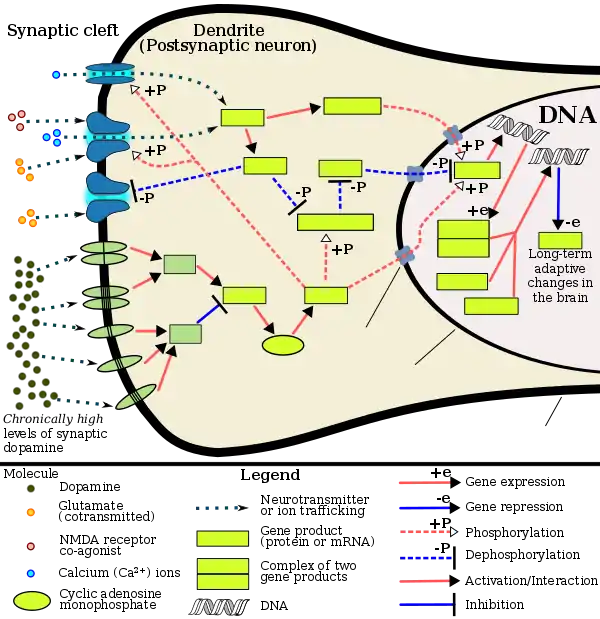
[Psychostimulants] increase cAMP levels in striatum, which activates protein kinase A (PKA) and leads to phosphorylation of its targets. This includes the cAMP response element binding protein (CREB), the phosphorylation of which induces its association with the histone acetyltransferase, CREB binding protein (CBP) to acetylate histones and facilitate gene activation. This is known to occur on many genes including fosB and c-fos in response to psychostimulant exposure. ΔFosB is also upregulated by chronic psychostimulant treatments, and is known to activate certain genes (eg, cdk5) and repress others (eg, c-fos) where it recruits HDAC1 as a corepressor. ... Chronic exposure to psychostimulants increases glutamatergic [signaling] from the prefrontal cortex to the NAc. Glutamatergic signaling elevates Ca2+ levels in NAc postsynaptic elements where it activates CaMK (calcium/calmodulin protein kinases) signaling, which, in addition to phosphorylating CREB, also phosphorylates HDAC5.
Figure 2: Psychostimulant-induced signaling events - ↑ Broussard JI (January 2012). "Co-transmission of dopamine and glutamate". The Journal of General Physiology. 139 (1): 93–96. doi:10.1085/jgp.201110659. PMC 3250102. PMID 22200950.
Coincident and convergent input often induces plasticity on a postsynaptic neuron. The NAc integrates processed information about the environment from basolateral amygdala, hippocampus, and prefrontal cortex (PFC), as well as projections from midbrain dopamine neurons. Previous studies have demonstrated how dopamine modulates this integrative process. For example, high frequency stimulation potentiates hippocampal inputs to the NAc while simultaneously depressing PFC synapses (Goto and Grace, 2005). The converse was also shown to be true; stimulation at PFC potentiates PFC–NAc synapses but depresses hippocampal–NAc synapses. In light of the new functional evidence of midbrain dopamine/glutamate co-transmission (references above), new experiments of NAc function will have to test whether midbrain glutamatergic inputs bias or filter either limbic or cortical inputs to guide goal-directed behavior.
- ↑ Kanehisa Laboratories (10 October 2014). "Amphetamine – Homo sapiens (human)". KEGG Pathway. Retrieved 31 October 2014.
Most addictive drugs increase extracellular concentrations of dopamine (DA) in nucleus accumbens (NAc) and medial prefrontal cortex (mPFC), projection areas of mesocorticolimbic DA neurons and key components of the "brain reward circuit". Amphetamine achieves this elevation in extracellular levels of DA by promoting efflux from synaptic terminals. ... Chronic exposure to amphetamine induces a unique transcription factor delta FosB, which plays an essential role in long-term adaptive changes in the brain.
- ↑ Cadet JL, Brannock C, Jayanthi S, Krasnova IN (2015). "Transcriptional and epigenetic substrates of methamphetamine addiction and withdrawal: evidence from a long-access self-administration model in the rat". Molecular Neurobiology. 51 (2): 696–717 (Figure 1). doi:10.1007/s12035-014-8776-8. PMC 4359351. PMID 24939695.
- 1 2 3 Robison AJ, Nestler EJ (November 2011). "Transcriptional and epigenetic mechanisms of addiction". Nature Reviews Neuroscience. 12 (11): 623–637. doi:10.1038/nrn3111. PMC 3272277. PMID 21989194.
ΔFosB serves as one of the master control proteins governing this structural plasticity. ... ΔFosB also represses G9a expression, leading to reduced repressive histone methylation at the cdk5 gene. The net result is gene activation and increased CDK5 expression. ... In contrast, ΔFosB binds to the c-fos gene and recruits several co-repressors, including HDAC1 (histone deacetylase 1) and SIRT 1 (sirtuin 1). ... The net result is c-fos gene repression.
Figure 4: Epigenetic basis of drug regulation of gene expression - 1 2 3 Nestler EJ (December 2012). "Transcriptional mechanisms of drug addiction". Clinical Psychopharmacology and Neuroscience. 10 (3): 136–143. doi:10.9758/cpn.2012.10.3.136. PMC 3569166. PMID 23430970.
The 35-37 kD ΔFosB isoforms accumulate with chronic drug exposure due to their extraordinarily long half-lives. ... As a result of its stability, the ΔFosB protein persists in neurons for at least several weeks after cessation of drug exposure. ... ΔFosB overexpression in nucleus accumbens induces NFκB ... In contrast, the ability of ΔFosB to repress the c-Fos gene occurs in concert with the recruitment of a histone deacetylase and presumably several other repressive proteins such as a repressive histone methyltransferase
- ↑ Nestler EJ (October 2008). "Transcriptional mechanisms of addiction: Role of ΔFosB". Philosophical Transactions of the Royal Society B: Biological Sciences. 363 (1507): 3245–3255. doi:10.1098/rstb.2008.0067. PMC 2607320. PMID 18640924.
Recent evidence has shown that ΔFosB also represses the c-fos gene that helps create the molecular switch—from the induction of several short-lived Fos family proteins after acute drug exposure to the predominant accumulation of ΔFosB after chronic drug exposure
- ↑ Mahner S, Baasch C, Schwarz J, Hein S, Wölber L, Jänicke F, Milde-Langosch K (October 2008). "C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma". Br. J. Cancer. 99 (8): 1269–75. doi:10.1038/sj.bjc.6604650. PMC 2570515. PMID 18854825.
- ↑ Graybiel AM, Moratalla R, Robertson HA (September 1990). "Amphetamine and cocaine induce drug-specific activation of the c-fos gene in striosome-matrix compartments and limbic subdivisions of the striatum". Proc. Natl. Acad. Sci. U.S.A. 87 (17): 6912–6. Bibcode:1990PNAS...87.6912G. doi:10.1073/pnas.87.17.6912. PMC 54648. PMID 2118661.
- ↑ Curran EJ, Akil H, Watson SJ (November 1996). "Psychomotor stimulant- and opiate-induced c-fos mRNA expression patterns in the rat forebrain: comparisons between acute drug treatment and a drug challenge in sensitized animals". Neurochem. Res. 21 (11): 1425–35. doi:10.1007/BF02532384. PMID 8947933. S2CID 6727581.
- ↑ Nichols CD, Sanders-Bush E (May 2002). "A single dose of lysergic acid diethylamide influences gene expression patterns within the mammalian brain". Neuropsychopharmacology. 26 (5): 634–42. doi:10.1016/S0893-133X(01)00405-5. PMID 11927188.
- 1 2 Singewald N, Salchner P, Sharp T (February 2003). "Induction of c-Fos expression in specific areas of the fear circuitry in rat forebrain by anxiogenic drugs". Biol. Psychiatry. 53 (4): 275–83. doi:10.1016/S0006-3223(02)01574-3. PMID 12586446. S2CID 29821546.
- ↑ VanElzakker M, Fevurly RD, Breindel T, Spencer RL (2008). "Environmental novelty is associated with a selective increase in c-fos expression in the output elements of the hippocampal formation and the perirhinal cortex". Learn. Mem. 15 (12): 899–908. doi:10.1101/lm.1196508. PMC 2632843. PMID 19050162.
- ↑ Dragunow M, Faull R (1989). "The use of c-fos as a metabolic marker in neuronal pathway tracing". Journal of Neuroscience Methods. 29 (3): 261–265. doi:10.1016/0165-0270(89)90150-7. PMID 2507830. S2CID 3804201.
- ↑ Day HE, Kryskow EM, Nyhuis TJ, Herlihy L, Campeau S (September 2008). "Conditioned Fear Inhibits c-fos mRNA Expression in the Central Extended Amygdala". Brain Res. 1229: 137–46. doi:10.1016/j.brainres.2008.06.085. PMC 2605076. PMID 18634767.
- ↑ Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, Bossert JM, Nair SG, Uejima JL, Marin MT, Mitchell TB, Farquhar D, Ghosh SC, Mattson BJ, Hope BT (August 2009). "Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization". Nat. Neurosci. 12 (8): 1069–73. doi:10.1038/nn.2364. PMC 2752202. PMID 19620976.
- ↑ Garner, Aleena (March 2012). "Generation of a Synthetic Memory Trace". Science. 335 (6075): 1513–1516. Bibcode:2012Sci...335.1513G. doi:10.1126/science.1214985. PMC 3956300. PMID 22442487.
- ↑ Na SY, Choi JE, Kim HJ, Jhun BH, Lee YC, Lee JW (October 1999). "Bcl3, an IkappaB protein, stimulates activating protein-1 transactivation and cellular proliferation". J. Biol. Chem. 274 (40): 28491–6. doi:10.1074/jbc.274.40.28491. PMID 10497212.
- ↑ Zhong H, Zhu J, Zhang H, Ding L, Sun Y, Huang C, Ye Q (December 2004). "COBRA1 inhibits AP-1 transcriptional activity in transfected cells". Biochem. Biophys. Res. Commun. 325 (2): 568–73. doi:10.1016/j.bbrc.2004.10.079. PMID 15530430.
- 1 2 Yamaguchi Y, Wada T, Suzuki F, Takagi T, Hasegawa J, Handa H (August 1998). "Casein kinase II interacts with the bZIP domains of several transcription factors". Nucleic Acids Res. 26 (16): 3854–61. doi:10.1093/nar/26.16.3854. PMC 147779. PMID 9685505.
- ↑ Ubeda M, Vallejo M, Habener JF (November 1999). "CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins". Mol. Cell. Biol. 19 (11): 7589–99. doi:10.1128/MCB.19.11.7589. PMC 84780. PMID 10523647.
- 1 2 Yang X, Chen Y, Gabuzda D (September 1999). "ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB". J. Biol. Chem. 274 (39): 27981–8. doi:10.1074/jbc.274.39.27981. PMID 10488148.
- ↑ Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H (January 2001). "Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers". J. Biol. Chem. 276 (4): 2852–7. doi:10.1074/jbc.M009633200. PMID 11053448.
- ↑ Pognonec P, Boulukos KE, Aperlo C, Fujimoto M, Ariga H, Nomoto A, Kato H (May 1997). "Cross-family interaction between the bHLHZip USF and bZip Fra1 proteins results in down-regulation of AP1 activity". Oncogene. 14 (17): 2091–8. doi:10.1038/sj.onc.1201046. PMID 9160889.
- ↑ Glover JN, Harrison SC (January 1995). "Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA". Nature. 373 (6511): 257–61. Bibcode:1995Natur.373..257G. doi:10.1038/373257a0. PMID 7816143. S2CID 4276971.
- ↑ Nomura N, Zu YL, Maekawa T, Tabata S, Akiyama T, Ishii S (February 1993). "Isolation and characterization of a novel member of the gene family encoding the cAMP response element-binding protein CRE-BP1". J. Biol. Chem. 268 (6): 4259–66. doi:10.1016/S0021-9258(18)53604-8. PMID 8440710.
- ↑ Finkel T, Duc J, Fearon ER, Dang CV, Tomaselli GF (January 1993). "Detection and modulation in vivo of helix-loop-helix protein-protein interactions". J. Biol. Chem. 268 (1): 5–8. doi:10.1016/S0021-9258(18)54105-3. PMID 8380166.
- ↑ Venugopal R, Jaiswal AK (December 1998). "Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes". Oncogene. 17 (24): 3145–56. doi:10.1038/sj.onc.1202237. PMID 9872330.
- ↑ Lee SK, Na SY, Jung SY, Choi JE, Jhun BH, Cheong J, Meltzer PS, Lee YC, Lee JW (June 2000). "Activating protein-1, nuclear factor-kappaB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2". Mol. Endocrinol. 14 (6): 915–25. doi:10.1210/mend.14.6.0471. PMID 10847592.
- ↑ Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, Lee JW (July 1998). "Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits". J. Biol. Chem. 273 (27): 16651–4. doi:10.1074/jbc.273.27.16651. PMID 9642216.
- ↑ Lee SK, Kim JH, Lee YC, Cheong J, Lee JW (April 2000). "Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor". J. Biol. Chem. 275 (17): 12470–4. doi:10.1074/jbc.275.17.12470. PMID 10777532.
- 1 2 Hess J, Porte D, Munz C, Angel P (June 2001). "AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element". J. Biol. Chem. 276 (23): 20029–38. doi:10.1074/jbc.M010601200. PMID 11274169.
- 1 2 D'Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC (January 2002). "Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation". J. Biol. Chem. 277 (1): 816–22. doi:10.1074/jbc.M107082200. PMID 11641401.
- ↑ Zhang Y, Feng XH, Derynck R (August 1998). "Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription". Nature. 394 (6696): 909–13. Bibcode:1998Natur.394..909Z. doi:10.1038/29814. PMID 9732876. S2CID 4393852.
- ↑ Metz R, Bannister AJ, Sutherland JA, Hagemeier C, O'Rourke EC, Cook A, Bravo R, Kouzarides T (September 1994). "c-Fos-induced activation of a TATA-box-containing promoter involves direct contact with TATA-box-binding protein". Mol. Cell. Biol. 14 (9): 6021–9. doi:10.1128/MCB.14.9.6021. PMC 359128. PMID 8065335.
- ↑ (Text color) Transcription factors
Further reading
- Murphy LC, Alkhalaf M, Dotzlaw H, Coutts A, Haddad-Alkhalaf B (June 1994). "Regulation of gene expression in T-47D human breast cancer cells by progestins and antiprogestins". Hum. Reprod. 9 Suppl 1: 174–80. doi:10.1093/humrep/9.suppl_1.174. PMID 7962462.
- Pompeiano M, Cirelli C, Arrighi P, Tononi G (1995). "c-Fos expression during wakefulness and sleep". Neurophysiol Clin. 25 (6): 329–41. doi:10.1016/0987-7053(96)84906-9. PMID 8904195. S2CID 23760149.
- Herrera DG, Robertson HA (October 1996). "Activation of c-fos in the brain". Prog. Neurobiol. 50 (2–3): 83–107. doi:10.1016/S0301-0082(96)00021-4. PMID 8971979. S2CID 31105978.
- Velazquez Torres A, Gariglio Vidal P (2002). "[Possible role of transcription factor AP1 in the tissue-specific regulation of human papillomavirus]". Rev. Invest. Clin. (in Spanish). 54 (3): 231–42. PMID 12183893.
External links
- c-fos+Proteins at the US National Library of Medicine Medical Subject Headings (MeSH)
- c-fos+Genes at the US National Library of Medicine Medical Subject Headings (MeSH)
- FactorBook c-Fos
- Drosophila kayak - The Interactive Fly
- Human FOS genome location and FOS gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P01100 (Human Proto-oncogene c-Fos) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P01101 (Mouse Proto-oncogene c-Fos) at the PDBe-KB.