2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase
In enzymology, a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) is an enzyme that catalyzes the chemical reaction
- 2,3-dihydro-2,3-dihydroxybenzoate + NAD+ 2,3-dihydroxybenzoate + NADH + H+
| 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 1.3.1.28 | ||||||||
| CAS no. | 37250-40-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Thus, the two substrates of this enzyme are 2,3-dihydro-2,3-dihydroxybenzoate and NAD+, whereas its 3 products are 2,3-dihydroxybenzoate, NADH, and H+.
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-CH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is 2,3-dihydro-2,3-dihydroxybenzoate:NAD+ oxidoreductase. This enzyme is also called 2,3-diDHB dehydrogenase. This enzyme participates in biosynthesis of siderophore group nonribosomal.[1]
Structure
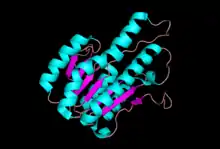
2,3-diDHB dehydrogenase is a tetramer protein with dimension 65x69x43 Å.[2] It has a crystallographic 222 symmetry, which exhibited for other members of short-chain oxireductase (SCOR) family of enzymes.[2] The length of each monomer is 248 residues and the weight of the protein is 24647 Da.[3] Each monomer consists of 7 beta-pleated sheets and 6 alpha helices.
Although the structure of the binding protein is not clearly defined, it was proposed that the binding pocket is made out of Leu83, Met85, Arg138, Gly140, Met141, Ser176, Met181, Gln182 and Leu185. It was also speculated that Arg138 is a likely subunit that interacts with the carboxyl group of 2,3-diDHB. Since there was a strong indication of oxidation at C3 position,[4] Ser176 and Gln182 interact with the C2-hydroxyl group in order for the stereo-selective reaction to occur.[2]
Reaction mechanism
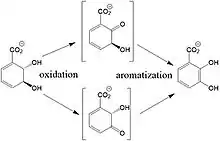
In times of limited iron number in the environment, the EntA reaction is irreversible physiologically.[4] The exact mechanism for the reaction is unknown; however, the proposed reaction scheme for the reaction is as following:
Rate of reaction
The rate of the conversion reaction is determined by several factors. The regiochemical position of the carboxyl group and the 3-hydroxyl group plays one role in the reaction, in which the rate of reaction of 1,3-cis-substituted substrate gives about 40-fold higher kcat/Km value than the 1,3-trans-substituted substrate.[4]
Roles in bacteria
E. coli
2,3-diDHB dehydrogenase catalyzes the NAD+-dependent oxidation of 2,3-dihydro-2,3-dihydroxybenzoate to produce an aromatic compound 2,3-dihydroxybenzoic acid (2,3-DHB or simply DHB).[4] In times of iron deficiency, iron uptake is controlled by three genes: ent, fep, and fes for synthesis, export, and uptake of ferric Enterobactin and its hydrolytic cleavage to release Fe3+ into the cell.[4] This production of this compound is controlled by eight genes: entA-entF,[2] entH,[5] and entS.[5] In E. coli, all of these genes are controlled by the Fur repressor, such that the genes are turned on when the concentration of iron in the environment is low.[5] From these six genes, EntA, EntB, and EntC are responsible for the synthesis of DHB from chorismic acid[2] and the gene EntA encodes the information of 2,3-diDHB dehydrogenase.[5] Without entA, entB, and entC, the bacteria show almost an absolute requirement of DHB in order to survive.[6]
A. tumefaciens
Production of siderophores also exhibited in some plant-infecting bacteria, such as Agrobacterium tumefaciens.[7] The enzyme is controlled by gene cluster agb and the production of 2,3-diDHB dehydrogenase is controlled by the gene agbA.[7] The enzyme AgbA is homologous to the EntA enzyme in E. coli, the same enzyme that produces 2,3-diDHB dehydrogenase.[7]
References
- Young IG, Gibson F (May 1969). "Regulation of the enzymes involved in the biosynthesis of 2,3-dihydroxybenzoic acid in Aerobacter aerogenes and Escherichia coli". Biochimica et Biophysica Acta (BBA) - General Subjects. 177 (3): 401–11. doi:10.1016/0304-4165(69)90302-X. PMID 4306838.
- Sundlov JA, Garringer JA, Carney JM, Reger AS, Drake EJ, Duax WL, Gulick AM (July 2006). "Determination of the crystal structure of EntA, a 2,3-dihydro-2,3-dihydroxybenzoic acid dehydrogenase from Escherichia coli". Acta Crystallographica D. 62 (Pt 7): 734–40. doi:10.1107/S0907444906015824. PMID 16790929.
- "Crystal Structure of E. coli EntA, a 2,3-dihydrodihydroxy benzoate dehydrogenase". RCSB Protein Data Bank. n.d. Retrieved 19 April 2015.
- Sakaitani M, Rusnak F, Quinn NR, Tu C, Frigo TB, Berchtold GA, Walsh CT (July 1990). "Mechanistic studies on trans-2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (Ent A) in the biosynthesis of the iron chelator enterobactin". Biochemistry. 29 (29): 6789–98. doi:10.1021/bi00481a006. PMID 2144454.
- Khalil S, Pawelek PD (February 2011). "Enzymatic adenylation of 2,3-dihydroxybenzoate is enhanced by a protein-protein interaction between Escherichia coli 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EntA) and 2,3-dihydroxybenzoate-AMP ligase (EntE)". Biochemistry. 50 (4): 533–45. doi:10.1021/bi101558v. PMID 21166461.
- Young IG, Langman L, Luke RK, Gibson F (April 1971). "Biosynthesis of the iron-transport compound enterochelin: mutants of Escherichia coli unable to synthesize 2,3-dihydroxybenzoate". Journal of Bacteriology. 106 (1): 51–7. doi:10.1128/JB.106.1.51-57.1971. PMC 248643. PMID 4928016.
- Sonoda H, Suzuki K, Yoshida K (June 2002). "Gene cluster for ferric iron uptake in Agrobacterium tumefaciens MAFF301001". Genes & Genetic Systems. 77 (3): 137–46. doi:10.1266/ggs.77.137. PMID 12207035.