ASUDAS
The ASUDAS (Arizona State University Dental Anthropology System) is a reference system for collecting data on human tooth morphology and variation created by Christy G. Turner II, Christian R. Nichol, and G. Richard Scott.[1] The ASUDAS gives detailed descriptions for common crown and root shape variants and their different degrees of expression. It also comprises a set of reference plaques illustrating dental variants as well as showing their expression levels in 3D. The ASUDAS was designed to ensure a standardized scoring procedure with minimum error in order to warrant comparability between data collected by different observers.
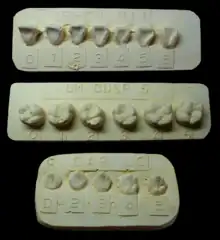
The ASUDAS currently comprises a set of 42 dental variants that can be observed in the permanent adult dentition.[2] The majority are crown and root shape variants, although the system also includes some skeletal variants of the maxilla and mandible. Most of the variants occur at different frequencies in human populations around the world.[3] Examples of dental variants listed in the ASUDAS are shovel-shaped incisors, Carabelli cusps, or hypocones.
Genetics
It is hypothesized that most of the dental variants listed in the ASUDAS are heritable and selectively neutral and that the worldwide dental diversity was generated by random evolutionary processes consisting of founder effects and genetic drift.[3] Several studies have also demonstrated that genetic distances across modern human populations derived from neutrally evolving SNPs or microsatellites are highly correlated with distances derived from dental variants listed in the ASUDAS.[4][5] Additionally, dental variation within populations decreases with increasing geographical distance from Africa,[6] a signature also found in neutral genetic datasets as a result of a serial founder effect originating in Africa.[7]
Some dental variants listed in the ASUDAS are also likely to be associated with non-neutral evolutionary processes, such as natural selection. For example, shoveling and double-shoveling of upper first incisors and the presence of hypoconulids of lower second molars have been found to be linked to the ectodysplasin A receptor gene (EDAR).[8][9] EDAR is a functional genomic region and has a range of pleiotropic effects on ectodermally derived structures, such as hair, mammary glands, and teeth, and is most likely under positive selection in Asian populations.[10][11] It is possible that dental variants linked to EDAR are not direct targets of positive selection but rather 'hitchhiking' when selection acts on another phenotype.[9]
Applications
The enamel which covers a tooth crown is the hardest tissue in the human body and generally well preserved in taphonomic contexts, even when associated skeletal and DNA preservation is relatively poor.[3] Therefore, dental morphological data collected with the ASUDAS are commonly used for inferring the biogeographical origin of deceased humans when no other biological markers are available. For example, ASUDAS data are typically used for identifying unknown individuals in forensic cases,[12] for examining past migration and mobility in bioarchaeological contexts,[13] and for reconstructing hominin phylogenies in paleoanthropological studies.[14]
As a rule of thumb, dental inferences about neutral genetic affinities based on many ASUDAS variants are more reliable than those based on only a few variants. Nevertheless, the best performance is achieved when using specific combinations of highly diagnostic variants, and not the full ASUDAS set.[15]
A study conducted in 2023 found that ASUDAS variants are among the most informative morphological markers in the human skeleton for inferring underlying neutral genetic relatedness among populations, significantly outperforming other commonly employed data types, such as cranial non-metric and dental metric variables, for instance.[16]
References
- Advances in dental anthropology. Kelley, Marc A., Larsen, Clark Spencer. New York: Wiley-Liss. 1991. pp. 13–32. ISBN 0-471-56839-2. OCLC 21599953.
{{cite book}}: CS1 maint: others (link) - Scott, G. Richard; Irish, Joel D. (2017). Human Tooth Crown and Root Morphology: The Arizona State University Dental Anthropology System. Cambridge: Cambridge University Press. doi:10.1017/9781316156629. ISBN 978-1-316-15662-9.
- Scott, G. Richard; Turner II, Christy G.; Townsend, Grant C.; Martinón-Torres, María (2018-03-15). The Anthropology of Modern Human Teeth: Dental Morphology and Its Variation in Recent and Fossil Homo sapiens (2 ed.). Cambridge University Press. doi:10.1017/9781316795859. ISBN 978-1-316-79585-9.
- Rathmann, Hannes; Reyes-Centeno, Hugo; Ghirotto, Silvia; Creanza, Nicole; Hanihara, Tsunehiko; Harvati, Katerina (2017-12-02). "Reconstructing human population history from dental phenotypes". Scientific Reports. 7 (1): 12495. Bibcode:2017NatSR...712495R. doi:10.1038/s41598-017-12621-y. ISSN 2045-2322. PMC 5624867. PMID 28970489.
- Irish, Joel D.; Morez, Adeline; Girdland Flink, Linus; Phillips, Emma L.W.; Scott, G. Richard (2020-04-01). "Do dental nonmetric traits actually work as proxies for neutral genomic data? Some answers from continental- and global-level analyses". American Journal of Physical Anthropology. 172 (3): 347–375. doi:10.1002/ajpa.24052. PMID 32237144.
- Hanihara, Tsunehiko (2008-02-06). "Morphological variation of major human populations based on nonmetric dental traits". American Journal of Physical Anthropology. 136 (2): 169–182. doi:10.1002/ajpa.20792. PMID 18257017.
- Ramachandran, S.; Deshpande, O.; Roseman, C. C.; Rosenberg, N. A.; Feldman, M. W.; Cavalli-Sforza, L. L. (2005-11-01). "Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa". Proceedings of the National Academy of Sciences. 102 (44): 15942–15947. Bibcode:2005PNAS..10215942R. doi:10.1073/pnas.0507611102. ISSN 0027-8424. PMC 1276087. PMID 16243969.
- Kimura, Ryosuke; Yamaguchi, Tetsutaro; Takeda, Mayako; Kondo, Osamu; Toma, Takashi; Haneji, Kuniaki; Hanihara, Tsunehiko; Matsukusa, Hirotaka; Kawamura, Shoji; Maki, Koutaro; Osawa, Motoki (2009-10-09). "A Common Variation in EDAR Is a Genetic Determinant of Shovel-Shaped Incisors". The American Journal of Human Genetics. 85 (4): 528–535. doi:10.1016/j.ajhg.2009.09.006. PMC 2756549. PMID 19804850.
- Park, Jeong-Heuy; Yamaguchi, Tetsutaro; Watanabe, Chiaki; Kawaguchi, Akira; Haneji, Kuniaki; Takeda, Mayako; Kim, Yong-Il; Tomoyasu, Yoko; Watanabe, Miyuki; Oota, Hiroki; Hanihara, Tsunehiko (2012-05-31). "Effects of an Asian-specific nonsynonymous EDAR variant on multiple dental traits". Journal of Human Genetics. 57 (8): 508–514. doi:10.1038/jhg.2012.60. ISSN 1434-5161. PMID 22648185.
- Bryk, Jarosław; Hardouin, Emilie; Pugach, Irina; Hughes, David; Strotmann, Rainer; Stoneking, Mark; Myles, Sean (2008-05-21). Stajich, Jason E. (ed.). "Positive Selection in East Asians for an EDAR Allele that Enhances NF-κB Activation". PLOS ONE. 3 (5): e2209. Bibcode:2008PLoSO...3.2209B. doi:10.1371/journal.pone.0002209. ISSN 1932-6203. PMC 2374902. PMID 18493316.
- Hlusko, Leslea J.; Carlson, Joshua P.; Chaplin, George; Elias, Scott A.; Hoffecker, John F.; Huffman, Michaela; Jablonski, Nina G.; Monson, Tesla A.; O’Rourke, Dennis H.; Pilloud, Marin A.; Scott, G. Richard (2018-05-08). "Environmental selection during the last ice age on the mother-to-infant transmission of vitamin D and fatty acids through breast milk". Proceedings of the National Academy of Sciences. 115 (19): E4426–E4432. doi:10.1073/pnas.1711788115. ISSN 0027-8424. PMC 5948952. PMID 29686092.
- Scott, G. Richard; Pilloud, Marin; Navega, David; Coelho, João; Cunha, Eugénia; Irish, Joel (January 2018). "rASUDAS: A New Web-Based Application for Estimating Ancestry from Tooth Morphology". Forensic Anthropology. 1 (1): 18–31. doi:10.5744/fa.2018.0003.
- Rathmann, Hannes; Kyle, Britney; Nikita, Efthymia; Harvati, Katerina; Saltini Semerari, Giulia (2019-11-14). "Population history of southern Italy during Greek colonization inferred from dental remains". American Journal of Physical Anthropology. 170 (4): 519–534. doi:10.1002/ajpa.23937. ISSN 0002-9483. PMID 31633202.
- Irish, J. D.; Guatelli-Steinberg, D.; Legge, S. S.; de Ruiter, D. J.; Berger, L. R. (2013-04-12). "Dental Morphology and the Phylogenetic "Place" of Australopithecus sediba". Science. 340 (6129): 1233062. doi:10.1126/science.1233062. ISSN 0036-8075. PMID 23580535. S2CID 206546794.
- Rathmann, Hannes; Reyes-Centeno, Hugo (May 19, 2020). "Testing the utility of dental morphological trait combinations for inferring human neutral genetic variation". Proceedings of the National Academy of Sciences of the United States of America. 117 (20): 10769–10777. doi:10.1073/pnas.1914330117. PMC 7245130. PMID 32376635.
- Rathmann, Hannes; Perretti, Silvia; Porcu, Valentina; Hanihara, Tsunehiko; Scott, G Richard; Irish, Joel D; Reyes-Centeno, Hugo; Ghirotto, Silvia; Harvati, Katerina (July 2023). "Inferring human neutral genetic variation from craniodental phenotypes". PNAS Nexus. 2 (7). doi:10.1093/pnasnexus/pgad217. ISSN 2752-6542. PMC 10338903. PMID 37457893.