ATP6V1G2
V-type proton ATPase subunit G 2 is an enzyme that in humans is encoded by the ATP6V1G2 gene.[5][6]
| ATP6V1G2 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ATP6V1G2, ATP6G, ATP6G2, NG38, VMA10, ATPase H+ transporting V1 subunit G2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 606853 MGI: 1913487 HomoloGene: 41518 GeneCards: ATP6V1G2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
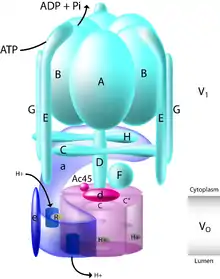
This gene encodes a component of vacuolar ATPase (V-ATPase), a multisubunit enzyme that mediates acidification of intracellular compartments of eukaryotic cells. V-ATPase dependent acidification is necessary for such intracellular processes as protein sorting, zymogen activation, receptor-mediated endocytosis, and synaptic vesicle proton gradient generation. V-ATPase is composed of a cytosolic V1 domain and a transmembrane V0 domain. The V1 domain consists of three A and three B subunits, two G subunits plus the C, D, E, F, and H subunits. The V1 domain contains the ATP catalytic site. The V0 domain consists of five different subunits: a, c, c', c double prime, and d.
Additional isoforms of many of the V1 and V0 subunit proteins are encoded by multiple genes, or alternatively spliced transcript variants. This encoded protein is one of three V1 domain G subunit proteins. This gene had previous gene symbols of ATP6G and ATP6G2. Alternatively spliced transcript variants encoding different isoforms have been described.[6]
Subcellular and tissue distribution
ATP6V1G2 is a subunit of a protein that has been identified in cell membranes, including intracellular membranes. These include lysosome, vacuole, and vesicle membranes within the cell.[7]
ATP6V1G2 is mostly found in the brain, and a smaller amount is found in the adrenals.[8] The enzyme is coded for by 4 exons.[8] The enzyme serves three main functions within the cell. First, ATP is hydrolyzed by this enzyme.[8] This means that ATP is broken down with water into ADP and a Hydrogen ion.[9] This Hydrogen ion serves as energy to drive other processes.[9] ATP6V1G2 also allows other proteins to bind within the cell.[8] ATP6V1G2 also allows ATPase to work by bringing Hydrogen ions into the cell.[8]
Structure
ATP6V1G2 is made of 118 amino acids.[10] ATP6V1G2 can make the pH of certain areas lower.[11]
Function
The biochemical abilities of ATP6V1G2 are involved in two other processes.[8] The first one includes managing autophagy within the cell.[8] The second is the decrease in pH of vesicles in the synapse.[8]
ATP6V1G2 is the specific part of the enzyme that hydrolyses ATP as a peripheral protein.[12]
There are approximately 45,000 ATP6V1G2 proteins in the membrane, with "150 proton pumps per" micrometers squared.[13]
ATP6V1G2 is a subunit of a protein.[13] It is in eukaryotic cells.[13] It pumps protons.[13] It is usually in internal plasma membranes.[13] The V1 portion of the protein "is an ATPase" and is outside of the plasma membrane.[13] ATP is required for hydrogens to enter the vesicle.[13] The V0 part of the protein is in the plasma membrane and the "a subunit" is used to determine the isoform of the protein.[13] Different isoforms are found in different tissues in mammals.[13] The release of V1 from V0 after protons have entered the vesicle and neurotransmitters, allows the V0 domain to travel with the vesicle to bind to another V0 domain and transfer the neurotransmitters.[13]
There is a set amount of G2 and G1 subunits of the protein.[14]
ATP6V1G2 has significant functions regarding the function of nerve signals.[13] The acidification of the interior of vesicles by ATP6V1G2 creates a difference in pH that is required for the neurotransmitters to enter the vesicle.[13] In this way, ATP6V1G2 allows for the preparation of neurotransmitters in the nerve vesicle.[13]
The decrease in pH by ATP6V1G2 in the vesicle is important in the function of vesicle binding to the SNARE protein and endocytosis.[13]
Activation of a nerve causes lower pH in the vesicles, and a larger pH in the cell.[13]
Calcium and hydrogen antiporters are required for ATP6V1G2 to acidify the inside of the vesicle.[13] The calcium is required to exit the cell in order to increase the number of hydrogens.[13] ATP6V1G2 lowers the proton concentration in the cell by the vesicle binding to the plasma membrane and lowering the concentration of the protons.[13]
The ATP6V1G2 functions to prepare the vesicle for neurotransmitters to enter the vesicle, the binding of the vesicle to the plasma membrane, and the endocytosis of the vesicle.[13]
vATPase helps in the process of neurotransmitters being brought into the vesicle and in the binding of the vesicle to the synapse.[14] The ATP6V1G2 part of the vATPase offers catalytic ability.[14] ATP6V1G2 is needed for function, however, no abnormalities were seen in its absence in the experiment of turning off the gene in mice.[14] ATP6V1G1 was increased when ATP6V1G2 was not present.[14] There was not more mRNA with the absence of ATP6V1G2.[14] More ATP6V1G1 was made without increased transcription.[14]
ATP6V1G2 completes processes involved with moving substances within the cell, as well as membranes, and the digestion of food.[14]
The functioning of vATPase is required for life.[14] The processes of vATPase allows for the immune cells to remove microorganisms, by the macrophage.[14] The vATPase also allows for T-cells and antigens to function.[14] The vATPase is also involved in acidifying the extracellular area of "bone resorbing osteoclasts," and "epithelial cells in the kidney."[14]
The ATP6V1G2 is the part of the protein that connects the V1 and V0 components of the protein together.[14] The ATP6V1G2 is required for the "energy coupling" between V1 and V0, allowing V1 and V0 to attach or unattach.[14] The G component of vATPase weighs 13 kDa.[14] ATP6V1G2 is found in the brain.[14] The experiment of creating a nonfunctional ATP6V1G2 gene in mice created no effects for offspring.[14] ATP6V1G2 is involved in ATP hydrolysis.[14] ATP6V1G2 was not able to substitute for Vma10 decrease, while ATP6V1G1 was.[14]
Clinical significance
Dysfunction of ATP6V1G2 leads to various disorders.[12] These can include "Noonan Syndrome 9," "Distal Renal Tubular Acidosis," and "Noonan Syndrome 3.".[12]
ATP6V1G2 may be involved in autoimmune diseases.[15] Activated macrophages created more ATP6V1G2.[15] The ATP6V1G2 gene is found in a 122kb group in the "TNF locus."[15] The increased ATP6V1G2 was independent of TNF.[15] The mRNA determines how much ATP6V1G2 is made because of an activated macrophage.[15] ATP6V1G2 may be a result of the inflammatory response.[15] ATP6V1G2 is involved with "protein sorting and degradation," "generation of secretory granules and endocytosis, and is known to be important for inflammatory and immune cell differentiation and function."[15] Factors like increased salt can increase vATPase.[15] Neutrophils, "degranulation," and "phagocytosis" from protein kinase C increases vATPase.[15] vATPase activity creates immune response against pathogens by macrophage activation.[15] Dendritic cells require vATPase to mature.[15]
References
- ENSG00000234920, ENSG00000206445, ENSG00000230900, ENSG00000213760, ENSG00000234668, ENSG00000226850 GRCh38: Ensembl release 89: ENSG00000227587, ENSG00000234920, ENSG00000206445, ENSG00000230900, ENSG00000213760, ENSG00000234668, ENSG00000226850 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000024403 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Neville MJ, Campbell RD (April 1999). "A new member of the Ig superfamily and a V-ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC". Journal of Immunology. 162 (8): 4745–4754. PMID 10202016.
- "Entrez Gene: ATP6V1G2 ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2".
- "Create Account". biocyc.org. Retrieved 2023-04-20.
- "ATP6V1G2 ATPase H+ transporting V1 subunit G2 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2023-04-20.
- "AmiGO 2: Term Details for "ATP hydrolysis activity" (GO:0016887)". Gene Ontology Consortium. Retrieved 2023-04-20.
- "26 items (human) - STRING interaction network". string-db.org. Retrieved 2023-04-25.
- "ATP6V1G2". www.nextprot.org. Retrieved 2023-04-25.
- "ATP6V1G2 Gene - GeneCards | VATG2 Protein | VATG2 Antibody". www.genecards.org. Retrieved 2023-04-25.
- Tabares L, Betz B (December 2010). "Multiple functions of the vesicular proton pump in nerve terminals". Neuron. 68 (6): 1020–1022. PMID 21172605.
- Kawamura N, Sun-Wada GH, Wada Y (September 2015). "Loss of G2 subunit of vacuolar-type proton transporting ATPase leads to G1 subunit upregulation in the brain". Scientific Reports. 5 (1): 14027. doi:10.1038/srep14027. PMC 4564858. PMID 26353914.
- Mewar D, Marinou I, Lee ME, Timms JM, Kilding R, Teare MD, et al. (December 2006). "Haplotype-specific gene expression profiles in a telomeric major histocompatibility complex gene cluster and susceptibility to autoimmune diseases". Genes and Immunity. 7 (8): 625–631. doi:10.1038/sj.gene.6364339. PMID 16971954.
Further reading
- Finbow ME, Harrison MA (June 1997). "The vacuolar H+-ATPase: a universal proton pump of eukaryotes". The Biochemical Journal. 324 ( Pt 3) (Pt 3): 697–712. doi:10.1042/bj3240697. PMC 1218484. PMID 9210392.
- Stevens TH, Forgac M (1998). "Structure, function and regulation of the vacuolar (H+)-ATPase". Annual Review of Cell and Developmental Biology. 13: 779–808. doi:10.1146/annurev.cellbio.13.1.779. PMID 9442887.
- Nelson N, Harvey WR (April 1999). "Vacuolar and plasma membrane proton-adenosinetriphosphatases". Physiological Reviews. 79 (2): 361–385. doi:10.1152/physrev.1999.79.2.361. PMID 10221984. S2CID 1477911.
- Forgac M (May 1999). "Structure and properties of the vacuolar (H+)-ATPases". The Journal of Biological Chemistry. 274 (19): 12951–12954. doi:10.1074/jbc.274.19.12951. PMID 10224039.
- Kane PM (February 1999). "Introduction: V-ATPases 1992-1998". Journal of Bioenergetics and Biomembranes. 31 (1): 3–5. doi:10.1023/A:1001884227654. PMID 10340843.
- Wieczorek H, Brown D, Grinstein S, Ehrenfeld J, Harvey WR (August 1999). "Animal plasma membrane energization by proton-motive V-ATPases". BioEssays. 21 (8): 637–648. doi:10.1002/(SICI)1521-1878(199908)21:8<637::AID-BIES3>3.0.CO;2-W. PMID 10440860. S2CID 23505139.
- Nishi T, Forgac M (February 2002). "The vacuolar (H+)-ATPases--nature's most versatile proton pumps". Nature Reviews. Molecular Cell Biology. 3 (2): 94–103. doi:10.1038/nrm729. PMID 11836511. S2CID 21122465.
- Kawasaki-Nishi S, Nishi T, Forgac M (June 2003). "Proton translocation driven by ATP hydrolysis in V-ATPases". FEBS Letters. 545 (1): 76–85. doi:10.1016/S0014-5793(03)00396-X. PMID 12788495. S2CID 10507213.
- Morel N (October 2003). "Neurotransmitter release: the dark side of the vacuolar-H+ATPase". Biology of the Cell. 95 (7): 453–457. doi:10.1016/S0248-4900(03)00075-3. PMID 14597263. S2CID 17519696.
- Smith AN, Borthwick KJ, Karet FE (September 2002). "Molecular cloning and characterization of novel tissue-specific isoforms of the human vacuolar H(+)-ATPase C, G and d subunits, and their evaluation in autosomal recessive distal renal tubular acidosis". Gene. 297 (1–2): 169–177. doi:10.1016/S0378-1119(02)00884-3. PMID 12384298.
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J (May 2003). "Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides". Nature Biotechnology. 21 (5): 566–569. doi:10.1038/nbt810. PMID 12665801. S2CID 23783563.
External links
- Human ATP6V1G2 genome location and ATP6V1G2 gene details page in the UCSC Genome Browser.