Adenylyl-sulfate reductase
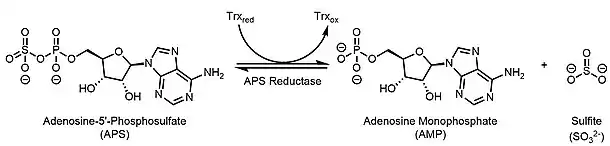
Adenylyl-sulfate reductase (EC 1.8.99.2) is an enzyme that catalyzes the chemical reaction of the reduction of adenylyl-sulfate/adenosine-5'-phosphosulfate (APS) to sulfite through the use of an electron donor cofactor. The products of the reaction are AMP and sulfite, as well as an oxidized electron donor cofactor.
| adenylyl-sulfate reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 adenylyl-sulfate reductase heterotetramer, Archaeoglobus fulgidus | |||||||||
| Identifiers | |||||||||
| EC no. | 1.8.99.2 | ||||||||
| CAS no. | 9027-75-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Nomenclature
This enzyme belongs to the family of oxidoreductases, specifically those acting on a sulfur group of donors with other acceptors. The systematic name of this enzyme class is AMP, sulfite:acceptor oxidoreductase (adenosine-5'-phosphosulfate-forming). Other names in common use include adenosine phosphosulfate reductase, adenosine 5'-phosphosulfate reductase, APS-reductase, APS reductase, AMP, sulfite:(acceptor) oxidoreductase, and (adenosine-5'-phosphosulfate-forming). This enzyme participates in selenium metabolism and sulfur metabolism.[1]
Mechanism
APS reductase catalyzes the reversible transformation of APS to sulfite and AMP, which is the rate determining step of the overall reaction.[2] The reaction catalyzed by APS reductase is as follows:
Sulfate has to be activated to APS by ATP sulfurylase at the expense of one ATP, hence this reaction requires an input of energy.[2] The reaction above occurs in a strictly anaerobic environment.[2] The two electrons come from a reduced cofactor, in this case reduced FAD.[3] The forward direction requires one AMP molecule; however, research suggests that the reverse reaction requires two AMP molecules (one acting on the substrate and one inhibiting the forward reaction).[3] The reversible reaction occurs when AMP binds to the Arg317 residue, changing the confirmation of Arg317 and APS reductase as a whole, which provides the thermodynamic driving force to go in the reverse direction.[3]
APS reductases are involved in both assimilatory and dissimilatory sulfate reduction.[4] Dissimilatory sulfate reduction takes sulfate and transforms it into sulfide, a sulfur source that can be distributed throughout the body.[4] Assimilatory sulfate reduction takes sulfate and turns it into cysteine.[4] Dissimilatory and assimilatory pathways both use APS reductases as a metabolic tool to produce a sulfur source and amino acids, respectively.[4]
Structure
As of late 2014, 6 structures have been solved for this class of enzymes, with PDB accession codes 1JNR, 1JNZ, 2FJA, 2FJB, 2FJD, and 2FJE.
The monomer of the enzyme consists of a mix of α-helices and β-sheets (both parallel and antiparallel). The protein cofactor thioredoxin can provide the required reducing equivalents for the reaction in the form of two cysteine residues, which are ultimately oxidized to a disulfide bond.[5] The base active form of APS reductase appears to be a heterodimer, as seen in plants.[6] In both bacteria and plants, two heterodimers tend to form together and produce a heterotetramer.[7]
The active site cleft in bacterial APS reductase has a few key elements. Residue sequences that appear to be necessary for catalysis are the P-loop (residues 60-66), the Arg-loop (residues 162-173), and the LDTG motif (residues 85-88). The P-loop, or phosphate-binding loop, is an especially important consecutive sequence of resides which aids in the recognition of the phosphate group in APS and, as a result, influences the substrate specificity for APS reductase. The C-terminal Cys256 is also catalytically essential, and seems to have a role in changing the conformation of the enzyme during catalysis.[5]
One notable chemical motif that distinguishes APS reductase from the related 3'-phosphoadenosine-5'-phosphosulfate (PAPS) reductase is the presence of a conserved cysteine motif, CC-X~80-CXXC, which occurs in addition to the universally conserved catalytic cysteine residue. This motif is correlated with the presence of a [4Fe-4S] cluster; therefore, these iron-sulfur clusters are not present within PAPS reductase. When the iron-sulfur cluster is present, it is required for catalytic activity and coordinated to the four cysteine residues in the conserved motif on the other side of the active site cleft.[5]
Function
Sulfur is a vital component in biological life and a key element in amino acids cysteine and methionine.[6] APS reductase controls the rate limiting step of endogenous sulfur assimilation, which is the process of producing hydrogen sulfide from sulfite. Hydrogen sulfite is one of the major sources of sulfur in plants.[6] APS reductase controls the flow of inorganic sulfur to cysteine, which is involved in many biological processes in plants such as growth, development, and responses to biotic and abiotic stresses.[6] In fact, studies have shown that when cells are starved of sulfur, APS reductase gene expression fluctuates, indicating that when the plants are exposed to metabolic and regulatory stress, APS reductase is likely a crucial enzyme in producing hydrogen sulfide and restoring homeostasis.[6]
Bacteria use APS reductases to engage in assimilatory and dissimilatory sulfate reduction, which make them prime candidates to appear in wastewater treatment environments.[8] Biofoulants can contain a number of sulfate reducing bacteria, and studies have shown that if wastewater plants are left untreated sulfate levels will decrease.[8] These studies have further solidified APS reductase’s crucial role in the global sulfur cycle by giving organisms another unique way to obtain sulfur when it's unavailable.[8]
Clinical significance
APS reductase does not exist within the proteome of human cells; consequently, these enzymes have become the targets of research for various environmental and medical reasons. Competitive inhibitors for the APS reductase in Mycobacterium tuberculosis have been studied as a new possible route for TB treatment, especially against drug-resistant and latent TB.[9] Such inhibitors have also been studied in the context of obtaining oil and gas from reservoirs in order to better control the souring of such products.[10]
Some APS reductases have also been investigated for their role in selenium metabolism and reduction due to the chemical similarity between sulfur and selenium. APR2, the dominant APS reductase isozyme in the model plant Arabidopsis thaliana, has been implicated in the involvement of selenate tolerance and selenite metabolism. Such research may then aid in the goal of enhancing selenium phytoremediation in plants and, as a result, dietary biofortification.[1]
References
- Grant K, Carey NM, Mendoza M, Schulze J, Pilon M, Pilon-Smits EA, van Hoewyk D (September 2011). "Adenosine 5'-phosphosulfate reductase (APR2) mutation in Arabidopsis implicates glutathione deficiency in selenate toxicity". The Biochemical Journal. 438 (2): 325–35. doi:10.1042/BJ20110025. PMID 21585336.
- Schiffer A, Fritz G, Kroneck PM, Ermler U (March 2006). "Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states". Biochemistry. 45 (9): 2960–7. doi:10.1021/bi0521689. PMID 16503650.
- Wójcik-Augustyn A, Johansson AJ, Borowski T (January 2021). "Reaction mechanism catalyzed by the dissimilatory adenosine 5'-phosphosulfate reductase. Adenosine 5'-monophosphate inhibitor and key role of arginine 317 in switching the course of catalysis". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1862 (1): 148333. doi:10.1016/j.bbabio.2020.148333. PMID 33130026. S2CID 226235547.
- Kushkevych I, Cejnar J, Treml J, Dordević D, Kollar P, Vítězová M (March 2020). "Recent Advances in Metabolic Pathways of Sulfate Reduction in Intestinal Bacteria". Cells. 9 (3): 698. doi:10.3390/cells9030698. PMC 7140700. PMID 32178484.
- Chartron J, Carroll KS, Shiau C, Gao H, Leary JA, Bertozzi CR, Stout CD (November 2006). "Substrate recognition, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine 5'-phosphosulfate reductase". Journal of Molecular Biology. 364 (2): 152–69. doi:10.1016/j.jmb.2006.08.080. PMC 1769331. PMID 17010373.
- Fu Y, Tang J, Yao GF, Huang ZQ, Li YH, Han Z, et al. (2018). "Central Role of Adenosine 5'-Phosphosulfate Reductase in the Control of Plant Hydrogen Sulfide Metabolism". Frontiers in Plant Science. 9: 1404. doi:10.3389/fpls.2018.01404. PMC 6166572. PMID 30319669.
- Fritz G, Roth A, Schiffer A, Büchert T, Bourenkov G, Bartunik HD, et al. (February 2002). "Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution". Proceedings of the National Academy of Sciences of the United States of America. 99 (4): 1836–1841. Bibcode:2002PNAS...99.1836F. doi:10.1073/pnas.042664399. PMC 122280. PMID 11842205.
- Zhou L, Ou P, Zhao B, Zhang W, Yu K, Xie K, Zhuang WQ (January 2021). "Assimilatory and dissimilatory sulfate reduction in the bacterial diversity of biofoulant from a full-scale biofilm-membrane bioreactor for textile wastewater treatment". The Science of the Total Environment. 772: 145464. Bibcode:2021ScTEn.772n5464Z. doi:10.1016/j.scitotenv.2021.145464. PMID 33571768. S2CID 231898490.
- Cosconati S, Hong JA, Novellino E, Carroll KS, Goodsell DS, Olson AJ (November 2008). "Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5'-phosphosulfate reductase inhibitors". Journal of Medicinal Chemistry. 51 (21): 6627–30. doi:10.1021/jm800571m. PMC 2639213. PMID 18855373.
- Dos Santos ES, de Souza LC, de Assis PN, Almeida PF, Ramos-de-Souza E (2014). "Novel potential inhibitors for adenylylsulfate reductase to control souring of water in oil industries". Journal of Biomolecular Structure & Dynamics. 32 (11): 1780–92. doi:10.1080/07391102.2013.834850. PMID 24028628. S2CID 31496404.
Further reading
- Michaels GB, Davidson JT, Peck HD (May 1970). "A flavin-sulfite adduct as an intermediate in the reaction catalyzed by adenylyl sulfate reductase from Desulfovibrio vulgaris". Biochemical and Biophysical Research Communications. 39 (3): 321–8. doi:10.1016/0006-291X(70)90579-6. PMID 5421934.