Anti-CRISPR
Anti-CRISPR (Anti-Clustered Regularly Interspaced Short Palindromic Repeats or Acr) is a group of proteins found in phages, that inhibit the normal activity of CRISPR-Cas, the immune system of certain bacteria.[1] CRISPR consists of genomic sequences that can be found in prokaryotic organisms, that come from bacteriophages that infected the bacteria beforehand, and are used to defend the cell from further viral attacks.[2] Anti-CRISPR results from an evolutionary process occurred in phages in order to avoid having their genomes destroyed by the prokaryotic cells that they will infect.[3]
| Anti-CRISPR (AcrIIA4 protein) | |||||||
|---|---|---|---|---|---|---|---|
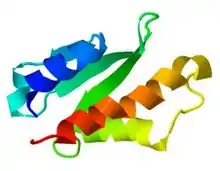 AcrIIA4 structure obtained from PDB with JSmol viewer. | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | AcrIIA4 | ||||||
| PDB | 5XN4 | ||||||
| UniProt | A0A247D711 | ||||||
| |||||||
Before the discovery of this type of family proteins, the acquisition of mutations was the only way known that phages could use to avoid CRISPR-Cas mediated shattering, by reducing the binding affinity of the phage and CRISPR. Nonetheless, bacteria have mechanisms to retarget the mutant bacteriophage, a process that it is called "priming adaptation". So, as far as researchers currently know, anti-CRISPR is the most effective way to ensure the survival of phages throughout the infection process of bacteria.[4]
History
Anti-CRISPR systems were first seen in Pseudomonas aeruginosa prophages,[5] which disabled type I-F CRISPR–Cas system, characteristic of some strains of these bacteria. After analysing the genomic sequences of these phages, genes codifying five different Anti-CRISPR proteins (also named Acrs) were discovered. Such proteins were AcrF1, AcrF2, AcrF3, AcrF4 and AcrF5. Research found none of these proteins disrupted the expression of Cas genes nor the assembling of CRISPR molecules, so it was thought that those type I-F proteins directly affected the CRISPR–Cas interference.[6]
Further investigation confirmed this hypothesis with the discovery of 4 other proteins (AcrE1, AcrE2, AcrE3 and AcrE4), which were shown to impede Pseudomonas aeruginosa’s CRISPR-Cas system.[7] Furthermore, the locus of the genes codifying these type I-E proteins was really close to the one responsible for the type I-F proteins expression in the same group of phages, leading to the conclusion that both types of proteins worked together.[8] However, these first nine proteins shared no common sequence motifs, which would have made easier the identification of new Anti-CRISPR protein families.
Later on, it was seen that phages that produced such proteins also encoded a putative transcriptional regulator named Aca 1 (anti-CRISPR associated 1) which was genetically located really close to the anti-CRISPR genes. This regulatory protein is supposed to be the responsible for the anti-CRISPR gene expression during the infectious cycle of the phage, therefore, both types of proteins (anti-CRISPR and Aca1) seem to work together as a single mechanism.[5]
After some studies, a similar amino-acid sequence to that of Aca1 was found, leading to the discovery of Aca2, a new family of Aca proteins. Aca2 also revealed the existence of five new groups of type I-F anti-CRISPR proteins due to their genomic proximity: AcrF6, AcrF7, AcrF8, AcrF9 and AcrF10. These proteins were not only present in Pseudomonas aeruginosa’s phages, as they also affected other cells of the Pseudomonadota (formerly Proteobacteria).[6]
Thanks to the use of bioinformatic tools, in 2016, AcrIIC1, AcrIIC2 and AcrIIC3 protein families were discovered in Neisseria meningitidis (which had been infected by phages previously). Such proteins were the first inhibitors of type II CRISPR–Cas to be found (concretely, they impeded II-C CRISPR–Cas9, the type of mechanism used in the genetic edition of human cells).[9] A year later, a study confirmed the presence of type II-A CRISPR–Cas9 inhibitors (AcrIIA1, AcrIIA2, AcrIIA3 and AcrIIA4) in Listeria monocytogenes (infected by bacteriophages which introduced the anti-CRISPR proteins). Two of those proteins (AcrIIA2 and AcrIIA4) were demonstrated to work properly against Streptococcus pyogenes type II-A defensive CRISPR system.
The result of all this research has been the discovery of 21 different Anti-CRISPR protein families, despite other inhibitors may exist due to the quick mutational process of phages. Thus, more research is needed to unravel the complexity of anti-CRISPR systems.
Types
Anti-CRISPR genes can be found in different parts of the phage DNA: in the capsid, the tail and at the extreme end. Moreover, it has been found that many MGEs have two or even three Acr genes in a single operon, which suggest that they could have been exchanged between MGEs.[10]
As all proteins, Acr family proteins are formed by the translation and transduction of the genes, and their classification is based on the type of CRISPR-Cas system they inhibit, due to the fact that each anti-CRISPR protein inhibits a specific CRISPR-Cas system. Although not many anti-CRISPR proteins have been discovered, these are the ones that have been found so far:
| Anti-CRISPR protein family | Characterized member | CRISPR system inhibited | Number of amino acids |
|---|---|---|---|
| AcrE1 | JBD5‑34 (Pseudomonas aeruginosa) | I‑E | 100 |
| AcrE2 | JBD88a‑32 (P. aeruginosa) | I‑E | 84 |
| AcrE3 | DMS3‑30 (P. aeruginosa) | I‑E | 68 |
| AcrE4 | D3112‑31 (P. aeruginosa) | I‑E | 52 |
| AcrF1 | JBD30‑35 (P. aeruginosa) | I‑F | 78 |
| AcrF2 | D3112‑30 (P. aeruginosa) | I‑F | 90 |
| AcrF3 | JBD5‑35 (P. aeruginosa) | I‑F | 139 |
| AcrF4 | JBD26‑37 (P. aeruginosa) | I‑F | 100 |
| AcrF5 | JBD5‑36 (P. aeruginosa) | I‑F | 79 |
| AcrF6 | AcrF6Pae (P. aeruginosa) | I‑E and I‑F | 100 |
| AcrF7 | AcrF7Pae (P. aeruginosa) | I‑F | 67 |
| AcrF8 | AcrF8ZF40 (Pectobacterium phage ZF40) | I‑F | 92 |
| AcrF9 | AcrF9Vpa (Vibrio parahaemolyticus) | I‑F | 68 |
| AcrF10 | AcrF10Sxi (Shewanella xiamenensis) | I‑F | 97 |
| AcrIIA1 | AcrIIA1Lmo (Listeria monocytogenes) | II‑A | 149 |
| AcrIIA2 | AcrIIA2Lmo (L. monocytogenes) | II‑A | 123 |
| AcrIIA3 | AcrIIA3Lmo (L. monocytogenes) | II‑A | 125 |
| AcrIIA4 | AcrIIA4Lmo (L. monocytogenes) | II‑A | 87 |
| AcrIIC1 | AcrIIC1Nme (Neisseria meningitidis) | II‑C | 85 |
| AcrIIC2 | AcrIIC2Nme (N. meningitidis) | II‑C | 123 |
| AcrIIC3 | AcrIIC3Nme (N. meningitidis) | II‑C | 116 |
So far, genes encoding anti-CRISPR proteins have been found in myophages, siphophages, putative conjugative elements and pathogenicity islands.
Attempts have been made to find common surrounding genetic features of anti-CRISPR genes, but without any success. Nevertheless, the presence of an aca gene just below anti-CRISPR genes has been observed.[10]
The first Acr protein families to be discovered were AcrF1, AcrF2, AcrF3, AcrF4 and AcrF5.[5] These inhibitors are mainly found in Pseudomonas phages, which are capable of infecting Pseudomonas aeruginosas possessing a type I‑F CRISPR–Cas system. Then, in another study, AcrE1, AcrE2, AcrE3 and AcrE4 protein families were found to also inhibit the type I‑F CRISPR–Cas in Pseudomonas aeruginosas.[7]
Later on, AcrF6, AcrF7, AcrF8, AcrF9 and AcrF10 protein families, which were also able to inhibit type I‑F CRISPR–Cas, were found to be very common in Pseudomonadota MGEs.[10]
The first inhibitors of a type II CRISPR–Cas system were then discovered: AcrIIC1, AcrIIC2 and AcrIIC3, that block the type II‑C CRISPR–Cas9 activity of Neisseria meningitidis.[9]
Finally, AcrIIA1, AcrIIA2, AcrIIA3 and AcrIIA4 were found. These protein families have the ability to inhibit the type II‑A CRISPR–Cas system of Listeria monocytogenes.[11]
As for the naming convention of Acr family proteins, it is established as follows: firstly, the type of system inhibited, then a numerical value referring to the protein family and finally the source of the specific anti-CRISPR protein. For example, AcrF9Vpa is active against the type I-F CRISPR–Cas system. It also was the ninth anti-CRISPR described for this system, and it is encoded in an integrated MGE in a Vibrio parahaemolyticus genome.
Structure
As exposed above, there is a wide spectrum of anti-CRISPR proteins, but few of these have been deeply studied. One of the most studied and well-defined Acrs is AcrIIA4, which inhibits Cas9, thus blocking the II-A CRISPR-Cas system of Streptococcus pyogenes.
AcrIIA4
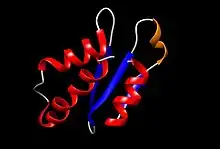
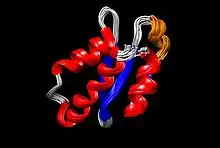 Structure of AcrIIA4 obtained with the UCSF Chimera software,[12] where its PDB file was uploaded.[13] Different colours were assigned to the four different secondary structures found in this protein: blue for β-strands, red for α-helices, orange for the 310 helix, and grey for loops. Originally, the PDB file contains the 20 lowest energy sequences (and thus, the most stable ones) superposed, one of which was randomly selected to create the figure.[14]
Structure of AcrIIA4 obtained with the UCSF Chimera software,[12] where its PDB file was uploaded.[13] Different colours were assigned to the four different secondary structures found in this protein: blue for β-strands, red for α-helices, orange for the 310 helix, and grey for loops. Originally, the PDB file contains the 20 lowest energy sequences (and thus, the most stable ones) superposed, one of which was randomly selected to create the figure.[14]The protein was solved using nuclear magnetic resonance (NMR); it contains 87 residues and its molecular weight is 10.182 kDa.[13] AcrIIA4 contains:
- 3 antiparallel β-strands (the first, from residues 16 to 19, the second, from 29 to 33, and the third, from 40 to 44) that form a β-sheet. This represents a 16,1% of the total number of amino acids, as 14 of them form the β-strands.
- 3 α-helices (the first, 2–13 residues, the second, 50–59 residues, and the third, 68–85 residues).
- 1 310 helix placed between the first (β1) and second (β2) β-strands, which starts at residue 22 and end in residue 25. The total helical part is composed of 40 residues, which is a 50,6% of the protein.
- Loops joining the different secondary structures.
There is a good definition of the secondary structures, as the three α-helices are packed near the three β-strands. Strikingly, between β3 strand, α2 and α3 helices, there is a hydrophobic core, originated by a cluster of aromatic side chains which are attracted by non-covalent interactions, such as pi stacking. Moreover, as it is an acidic protein, there is a high concentration of negatively charged residues in the loops between β3 and α2, between α2 and α3, and in the first part of α3, which may play an important role in the inhibition of Cas9, as negative charges might imitate phosphates of nucleic acids.[14]
AcrF1
On the other hand, there is another Acr, AcrF1, which may not have been as studied as the explained above, although there is a good description of its structure. It inhibits the I-F CRISPR-Cas system of Pseudomonas aeruginosa. Maxwell et al.[15] solved the 3D structure using NMR.
The protein contains 78 residues,[6] between which interact to form secondary structures. The structure of AcrF1 is formed of two anti-parallel α-helices and a β-sheet, which contains four anti-parallel β-strands. This β-sheet is placed in the contrary side of the α-helical part, which creates a hydrophobic core formed of 13 amino acids. Turns can also be found in different parts of the protein, for instance, joining the β-strands.[15][16]
There are surface residues which actively participate in the active site of AcrF1, two of which are tyrosines (Y6 and Y20) and the third amino acid is a glutamic acid (E31), as their mutation by an alanine causes a 100-fold decrease in the activity of the protein (with Y20A and E31A mutations), and a 107-fold decrease when Y6 is mutated.
The different structures that form the protein create a strange combination, as Maxwell et al. conducted a DALI search in order to find similarities between other proteins, and they found no informative similarities.[15]
Function
Avoiding destruction of the phage DNA
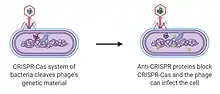
The principal function of anti-CRISPR proteins is to interact with specific components of CRISPR-Cas systems, such as the effector nucleases, to avoid the destruction of the phage DNA (by binding or cleavage).[17]
A phage introduces its DNA into a prokaryotic cell, usually the cell detects a sequence known as "target", that activates CRISPR-Cas immune system, but the presence of an initial sequence (before the target) encoding the formation of Acr proteins, avoids phage destruction. Acr proteins are formed before the target sequence is read. This way, the CRISPR-Cas system is blocked before it can develop a response.
The procedure starts with the CRISPR locus being transcribed into crRNAs (CRISPR RNA). CrRNAs combine with Cas proteins forming a ribonucleoprotein complex called Cascade. This complex surveys the cell to find complementary sequences of the crRNA. When this sequence is found, the Cas3 nuclease is recruited to the Cascade, and the target DNA from the phage is cleaved. But, for instance, when AcrF1 and AcrF2 are found (anti-CRISPR proteins), these interact with Cas7f and Cas8f-Cas5f, respectively, not allowing the binding to the phage DNA. Moreover, the cleaving of the target is prevented by the union between AcrF3 and Cas3.[6]
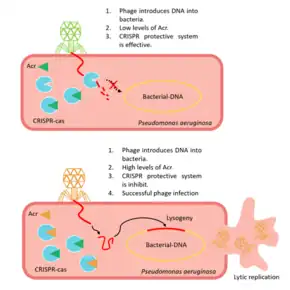
The majority of Acr genes are located next to anti-CRISPR-associated (Aca) genes, which encode proteins with a helix-turn-helix DNA-binding motif. Aca genes are preserved, and researchers are using them to identify Acr genes, but the function of the proteins they encode is not totally clear. The Acr-associated promoter produces high levels of Acr transcription just after the phage DNA injection into the bacteria takes place and, afterward, Aca proteins repress the transcription. If this wasn't repressed, the constant transcription of the gene would be lethal to the phage. Therefore, Aca activity is essential to ensure its survival.[18]
Phage-phage cooperation
Moreover, it has been verified that bacteria with CRISPR-Cas systems are still partially immune to Acr. Consequently, initial abortive phage infections may be unable to hamper CRISPR immunity, but phage-phage cooperation can increasingly boost Acr production and promote immunosuppression, which might produce an increase on the vulnerability of the host cell to reinfection, and finally allow a successful infection and spreading of a second phage.[17] This cooperation creates an epidemiological tipping point, in which, depending on the initial density of Acr-phages and the strength of CRISPR/Acr binding, phages can either be eliminated or originate a phage epidemic (the number of bacteriophages is amplified).[19][20]
If the starting levels of phages are high enough, the density of immunosuppressed hosts reaches a critical point where there are more successful infections than unsuccessful ones. Then, an epidemic begins. If this point is not reached, phage extinction occurs, and immunosuppressed hosts recover their initial state.[19][20]
Phage immune evasion
It has become clear that Acr proteins play an important role in allowing phage immune evasion, though it is still unclear how anti-CRISPR proteins synthesis can overcome the host’s CRISPR-Cas system, which can shatter the phage genome within minutes after the infection.[17]
Mechanisms
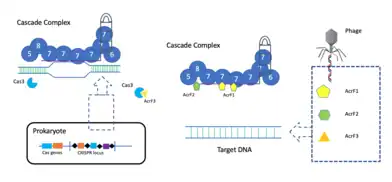
Within all the Anti-CRISPR proteins that have been discovered so far, mechanisms have been described for only 15 of among them. These mechanisms can be divided into three different types: crRNA loading interference, DNA binding blockage and DNA cleavage prevention.
CrRNA loading interference
CrRNA (CRISPR RNA) loading interference mechanism has been mainly associated with the AcrIIC2 protein family.[22] In order to block Cas9 activity, it prevents the correct assembly of the crRNA‐Cas9 complex.
DNA binding blockage
AcrIIC2 has been shown not to be the only one capable of blocking DNA binding. There are 11 other Acr family proteins that can also carry it out. Some among those are AcrIF1, AcrIF2, and AcrIF10, which act on different subunits of the Cascade effector complex of the type I‐F CRISPR‐Cas system, preventing the DNA to bind to the complex.[23]
Furthermore, AcrIIC3 prevents DNA binding by promoting dimerization of Cas9 [22][24] and AcrIIA2 mimics DNA, thereby blocking the PAM recognition residues and consequently preventing dsDNA (double-stranded DNA) recognition and binding.[25][26]
DNA cleavage prevention
AcrE1, AcrIF3 and AcrIIC1 can prevent target DNA cleavage. Using X-ray crystallography, AcrE1 was discovered to bind to the CRISPR associated Cas3.[27] Likewise, biochemical and structural analysis of AcrIF3 showed its capacity of binding to Cas3 as a dimer so as to prevent the recruitment of Cas3 to the Cascade complex.[23][28][29] Finally, thanks to biochemical and structural AcrIIC1 studies, it was found that it binds to the active site of the HNH endonuclease domain in Cas9, which prevents DNA from cleaving. Thus, it turns Cas9 into an inactive but DNA bound state.[24]
Applications
Reducing CRISPR-Cas9 off-target cuts
AcrIIA4 is one of the proteins responsible for the CRISPR-Cas9 system inhibition, the mechanism used in mammalian cells edition. Addition of AcrIIA4 in human cells avoids Cas9 interaction with the CRISPR system, reducing its ability to cut DNA. However, diverse studies have reached the conclusion that adding it in small proportions after the genome editing has been done, reduces the number of off-target cuts at the concrete sites in which Cas9 interacts, a thing that makes the whole system much more precise.[25]
Avoiding ecological consequences
One of the main objectives of using CRISPR-Cas9 technology is eradicating diseases, some of which are found in disease vectors, such as mosquitoes. Anti-CRISPR proteins can impede gene drive, which could create uncertain and catastrophic consequences in ecosystems.[30]
Detect presence of Cas9 in a sample
In order to know whether a certain bacterium synthesises Cas9, and therefore uses CRISPR-Cas9, or to detect accidental or not allowed use of this system, AcrIIC1 can be used. As the aforementioned protein binds to Cas9, a centrifugal microfluidic platform has been designed to detect it and determine its catalytic activity.[30]
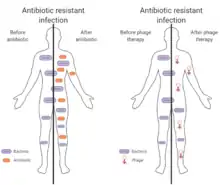
Phage therapy
Antibiotic resistance is a public health problem that is constantly increasing, because of the bad use of antibiotics. Phage therapy consists of the infection of bacteria using phages, which are much more specific and cause less side effects than antibiotics. Acrs could inhibit the CRISPR-Cas9 system of some bacteria and allow these phages to infect bacterial cells without being attacked by its immune system.[30]
See also
References
- Nakamura M, Srinivasan P, Chavez M, Carter MA, Dominguez AA, La Russa M, et al. (January 2019). "Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells". Nature Communications. 10 (1): 194. Bibcode:2019NatCo..10..194N. doi:10.1038/s41467-018-08158-x. PMC 6331597. PMID 30643127.
- Barrangou R (February 2015). "The roles of CRISPR-Cas systems in adaptive immunity and beyond". Current Opinion in Immunology. 32: 36–41. doi:10.1016/j.coi.2014.12.008. PMID 25574773.
- Stanley SY, Borges AL, Chen KH, Swaney DL, Krogan NJ, Bondy-Denomy J, Davidson AR (September 2019). "Anti-CRISPR-Associated Proteins Are Crucial Repressors of Anti-CRISPR Transcription". Cell. 178 (6): 1452–1464.e13. doi:10.1016/j.cell.2019.07.046. PMC 6754177. PMID 31474367.
- Maxwell KL (October 2017). "The Anti-CRISPR Story: A Battle for Survival". Molecular Cell. 68 (1): 8–14. doi:10.1016/j.molcel.2017.09.002. PMID 28985512.
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR (January 2013). "Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system". Nature. 493 (7432): 429–32. Bibcode:2013Natur.493..429B. doi:10.1038/nature11723. PMC 4931913. PMID 23242138.
- Pawluk A, Davidson AR, Maxwell KL (January 2018). "Anti-CRISPR: discovery, mechanism and function". Nature Reviews. Microbiology. 16 (1): 12–17. doi:10.1038/nrmicro.2017.120. PMID 29062071. S2CID 13222384.
- Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR (April 2014). "A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa". mBio. 5 (2): e00896. doi:10.1128/mBio.00896-14. PMC 3993853. PMID 24736222.
- Borges AL, Davidson AR, Bondy-Denomy J (September 2017). "The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs". Annual Review of Virology. 4 (1): 37–59. doi:10.1146/annurev-virology-101416-041616. PMC 6039114. PMID 28749735.
- Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, et al. (December 2016). "Naturally Occurring Off-Switches for CRISPR-Cas9". Cell. 167 (7): 1829–1838.e9. doi:10.1016/j.cell.2016.11.017. PMC 5757841. PMID 27984730.
- Pawluk A, Staals RH, Taylor C, Watson BN, Saha S, Fineran PC, et al. (June 2016). "Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species". Nature Microbiology. 1 (8): 16085. doi:10.1038/nmicrobiol.2016.85. PMID 27573108. S2CID 3826582.
- Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J (January 2017). "Inhibition of CRISPR-Cas9 with Bacteriophage Proteins". Cell. 168 (1–2): 150–158.e10. doi:10.1016/j.cell.2016.12.009. PMC 5235966. PMID 28041849.
- "UCSF Chimera". Chimera. Retrieved 25 October 2019.
- "AcrIIA4 - PDB". Protein Data Bank. doi:10.2210/pdb5xn4/pdb. Retrieved 2019-10-15.
- Kim I, Jeong M, Ka D, Han M, Kim NK, Bae E, Suh JY (March 2018). "Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor". Scientific Reports. 8 (1): 3883. Bibcode:2018NatSR...8.3883K. doi:10.1038/s41598-018-22177-0. PMC 5832863. PMID 29497118.
- Maxwell KL, Garcia B, Bondy-Denomy J, Bona D, Hidalgo-Reyes Y, Davidson AR (October 2016). "The solution structure of an anti-CRISPR protein". Nature Communications. 7 (1): 13134. Bibcode:2016NatCo...713134M. doi:10.1038/ncomms13134. PMC 5062604. PMID 27725669.
- Davidson AR, Pawluk A, Maxwell KL, Bondy-Denomy J. "AcrF1 - PDB". Worldwide Protein Data Bank. doi:10.2210/pdb2lw5/pdb. Retrieved 2019-10-14.
- van Gent M, Gack MU (September 2018). "Viral Anti-CRISPR Tactics: No Success without Sacrifice". Immunity. 49 (3): 391–393. doi:10.1016/j.immuni.2018.08.023. PMID 30231980.
- "Inici sessió - Identificació UB - Universitat de Barcelona". sso.ub.edu. Retrieved 2019-10-25.
- Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, et al. (August 2018). "Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity". Cell. 174 (4): 908–916.e12. doi:10.1016/j.cell.2018.05.058. PMC 6086933. PMID 30033365.
- Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, Bondy-Denomy J (August 2018). "Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity". Cell. 174 (4): 917–925.e10. doi:10.1016/j.cell.2018.06.013. PMC 6086726. PMID 30033364.
- Zhu Y, Zhang F, Huang Z (March 2018). "Structural insights into the inactivation of CRISPR-Cas systems by diverse anti-CRISPR proteins". BMC Biology. 16 (1): 32. doi:10.1186/s12915-018-0504-9. PMC 5859409. PMID 29554913.
- Zhu Y, Gao A, Zhan Q, Wang Y, Feng H, Liu S, et al. (April 2019). "Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins". Molecular Cell. 74 (2): 296–309.e7. doi:10.1016/j.molcel.2019.01.038. PMC 6750902. PMID 30850331.
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, et al. (October 2015). "Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins". Nature. 526 (7571): 136–9. Bibcode:2015Natur.526..136B. doi:10.1038/nature15254. PMC 4935067. PMID 26416740.
- Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ, Edraki A, et al. (September 2017). "A Broad-Spectrum Inhibitor of CRISPR-Cas9". Cell. 170 (6): 1224–1233.e15. doi:10.1016/j.cell.2017.07.037. PMC 5875921. PMID 28844692.
- Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, et al. (July 2017). "Disabling Cas9 by an anti-CRISPR DNA mimic". Science Advances. 3 (7): e1701620. Bibcode:2017SciA....3E1620S. doi:10.1126/sciadv.1701620. PMC 5507636. PMID 28706995.
- Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, et al. (June 2017). "Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein". Nature. 546 (7658): 436–439. Bibcode:2017Natur.546..436D. doi:10.1038/nature22377. PMID 28448066. S2CID 4445217.
- Pawluk A, Shah M, Mejdani M, Calmettes C, Moraes TF, Davidson AR, Maxwell KL (December 2017). "Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein". mBio. 8 (6). doi:10.1128/mBio.01751-17. PMC 5727412. PMID 29233895.
- Wang J, Ma J, Cheng Z, Meng X, You L, Wang M, et al. (September 2016). "A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses". Cell Research. 26 (10): 1165–1168. doi:10.1038/cr.2016.103. PMC 5113301. PMID 27585537.
- Wang X, Yao D, Xu JG, Li AR, Xu J, Fu P, et al. (September 2016). "Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3". Nature Structural & Molecular Biology. 23 (9): 868–70. doi:10.1038/nsmb.3269. PMID 27455460. S2CID 6466590.
- Zhang F, Song G, Tian Y (June 2019). "Anti-CRISPRs: The natural inhibitors for CRISPR-Cas systems". Animal Models and Experimental Medicine. 2 (2): 69–75. doi:10.1002/ame2.12069. PMC 6600654. PMID 31392299.