Auxotrophy
Auxotrophy (Ancient Greek: αὐξάνω "to increase"; τροφή "nourishment") is the inability of an organism to synthesize a particular organic compound required for its growth (as defined by IUPAC). An auxotroph is an organism that displays this characteristic; auxotrophic is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth.

Prototrophic cells (also referred to as the 'wild type') are self sufficient producers of all required metabolites (e.g. amino acids, lipids, cofactors), while auxotrophs require to be on medium with the metabolite that they cannot produce.[1] For example saying a cell is methionine auxotrophic means that it would need to be on a medium containing methionine or else it would not be able to replicate. In this example this is because it is unable to produce its own methionine (methionine auxotroph). However, a prototroph or a methionine prototrophic cell would be able to function and replicate on a medium with or without methionine.[2]
Replica plating is a technique that transfers colonies from one plate to another in the same spot as the last plate so the different media plates can be compared side by side. It is used to compare the growth of the same colonies on different plates of media to determine which environments the bacterial colony can or cannot grow in (this gives insight to possible auxotrophic characteristics. The method of replica plating implemented by Joshua Lederberg and Esther Lederberg included auxotrophs that were temperature-sensitive; that is, their ability to synthesize was temperature-dependent.[3] (Auxotrophs are usually not temperature-dependent. They can also depend on other factors.) It is also possible that an organism is auxotrophic to more than just one organic compound that it requires for growth.[4]
Applications
Genetics

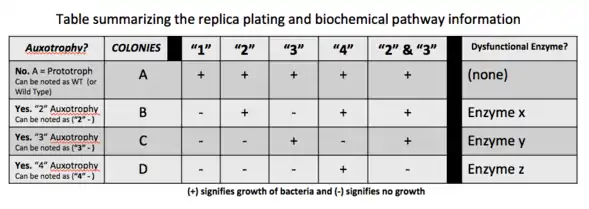
In genetics, a strain is said to be auxotrophic if it carries a mutation that renders it unable to synthesize an essential compound. For example, a yeast mutant with an inactivated uracil synthesis pathway gene is a uracil auxotroph (e.g., if the yeast Orotidine 5'-phosphate decarboxylase gene is inactivated, the resultant strain is a uracil auxotroph). Such a strain is unable to synthesize uracil and will only be able to grow if uracil can be taken up from the environment. This is the opposite of a uracil prototroph, or in this case a wild-type strain, which can still grow in the absence of uracil. Auxotrophic genetic markers are often used in molecular genetics; they were famously used in Beadle and Tatum's Nobel prize-winning work on the one gene-one enzyme hypothesis, connecting mutations of genes to protein mutations. This then allows for biosynthetic or biochemical pathway mapping that can help determine which enzyme or enzymes are mutated and dysfunctional in the auxotrophic strains of bacteria being studied.[2]
Researchers have used strains of E. coli auxotrophic for specific amino acids to introduce non-natural amino acid analogues into proteins. For instance cells auxotrophic for the amino acid phenylalanine can be grown in media supplemented with an analogue such as para-azido phenylalanine.
Many living things, including humans, are auxotrophic for large classes of compounds required for growth and must obtain these compounds through diet (see vitamin, essential nutrient, essential amino acid, essential fatty acid).
The complex pattern of evolution of vitamin auxotrophy across the eukaryotic tree of life is intimately connected with the interdependence between organisms.[5]

The Mutagenicity test (or Ames test)
The Salmonella Mutagenesis test (Ames test) uses multiple strains of Salmonella typhimurium that are auxotrophic to histidine to test whether a given chemical can cause mutations by observing its auxotrophic property in response to an added chemical compound.[6] The mutation a chemical substance or compound causes is measured by applying it to the bacteria on a plate containing histidine then moving the bacteria to a new plate without sufficient histidine for continual growth. If the substance does not mutate the genome of the bacteria from auxotrophic to histidine back to prototrophic to histidine, then the bacteria would not show growth on the new plate. So by comparing the ratio of the bacteria on the new plate to the old plate and the same ratio for the control group, it is possible to quantify how mutagenic a substance is, or rather, how likely it is to cause mutations in DNA.[7] A chemical is considered positive for Ames test if it causes mutations increasing the observed reversion rate and negative if presents similar to the control group. There is a normal, but small, number of revertant colonies expected when an auxotrophic bacteria is plated on a media without the metabolite it needs because it could mutate back to prototrophy. The chances of this are low and therefore cause very small colonies to be formed. If a mutagenic substance is added, however, the number of revertants would be visibly higher than without the mutagenic substance. The Ames test, basically, is considered positive if a substance increases chance of mutation in the DNA of the bacteria enough to cause a quantifiable difference in the revertants of the mutagen plate and the control group plate. Negative Ames test means the possible mutagen DID not cause increase in revertants and positive Ames test signifies that the possible mutagen DID increase the chance of mutation. These mutagenic effects on bacteria are researched as a possible indicator of the same effects on larger organisms, like humans. It is suggested that if a mutation can arise in bacterial DNA under presence of a mutagen then the same effect would occur for larger organisms causing cancer.[6] A negative Ames test result could suggest that the substance is not a mutagen and would not cause tumor formation in living organisms. However only few of the positive Ames Test resulting chemicals were considered insignificant when tested in larger organisms but the positive Ames test for bacteria still could not be conclusively linked to expression of cancer in larger organisms. While it can be a possible determinant of tumors for living organisms, humans, animals, and so on, more studies must be completed to come to a conclusion.[8]
Auxotrophy-based methods to incorporate unnatural amino acids into proteins and proteomes
A large number of unnatural amino acids, which are similar to their canonical counterparts in shape, size and chemical properties, are introduced into the recombinant proteins by means of auxotrophic expression hosts.[9] For example, methionine (Met) or tryptophan (Trp) auxotrophic Escherichia coli strains can be cultivated in a defined minimal medium. In this experimental setup it is possible to express recombinant proteins whose canonical Trp and Met residues are completely substituted with different medium-supplemented related analogs.[10] This methodology leads to a new form of protein engineering, which is not performed by codon manipulation at the DNA level (e.g. oligonucleotide-directed mutagenesis), but by codon reassignments at the level of protein translation under efficient selective pressure.[11] Therefore, the method is referred as selective pressure incorporation (SPI).[12]
No organism studied so far encodes other amino acids than the canonical twenty; two additional canonical amino acids (selenocysteine, pyrrolysine) are inserted into proteins by recoding translation termination signals. This boundary can be crossed by adaptive laboratory evolution of metabolically stable auxotrophic microbial strains. For example, the first clearly successful attempt to evolve Escherichia coli that can survive solely on the unnatural amino acid thieno[3,2-b]pyrrolyl) alanine as the only substitute for tryptophan was made in 2015.[13]
In popular culture
The 1993 film Jurassic Park (based on the 1990 Michael Crichton novel of the same name) features dinosaurs that were genetically altered so that they could not produce the amino acid lysine.[14] This was known as the "lysine contingency" and was supposed to prevent the cloned dinosaurs from surviving outside the park, forcing them to be dependent on lysine supplements provided by the park's veterinary staff. In reality, no animals are capable of producing lysine (it is an essential amino acid).[15]
See also
Footnotes
- Genetics : from genes to genomes. Hartwell, Leland. (4th ed.). New York: McGraw-Hill. 2011. ISBN 9780073525266. OCLC 317623365.
{{cite book}}: CS1 maint: others (link) - LaRossa, R.A. (2001). "Nutritional Mutations". Encyclopedia of Genetics. pp. 1362–1363. doi:10.1006/rwgn.2001.0920. ISBN 9780122270802.
- Lederberg, Joshua; Lederberg, Esther M. (March 1952). "Replica plating and indirect selection of bacterial mutants". Journal of Bacteriology. 63 (3): 399–406. doi:10.1128/JB.63.3.399-406.1952. ISSN 0021-9193. PMC 169282. PMID 14927572.
- Griffiths, Anthony J.F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (2000). "Mutant types".
{{cite journal}}: Cite journal requires|journal=(help) - Helliwell, Katherine E.; et al. (2013). "Widespread decay of vitamin-related pathways: coincidence or consequence?". Trends in Genetics. 29 (8): 469–478. doi:10.1016/j.tig.2013.03.003. PMID 23623319.
- Ahern, Kevin (2017). Biochemistry Free for All. DavinciPress.
- Ames, Bruce N.; McCann, Joyce; Yamasaki, Edith (1975). "Methods for detecting carcinogens and mutagens with the salmonella/mammalian-microsome mutagenicity test". Mutation Research/Environmental Mutagenesis and Related Subjects. 31 (6): 347–363. doi:10.1016/0165-1161(75)90046-1. PMID 768755.
- Kirkland, David; Zeiger, Errol; Madia, Federica; Gooderham, Nigel; Kasper, Peter; Lynch, Anthony; Morita, Takeshi; Ouedraogo, Gladys; Morte, Juan Manuel Parra (2014). "Can in vitro mammalian cell genotoxicity test results be used to complement positive results in the Ames test and help predict carcinogenic or in vivo genotoxic activity? I. Reports of individual databases presented at an EURL ECVAM Workshop". Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 775–776: 55–68. doi:10.1016/j.mrgentox.2014.10.005. PMID 25435356.
- Link, James; Mock, Marissa; Tirrell, David (2003). "Non-canonical amino acids in protein engineering". Curr. Opin. Biotechnol. 14 (6): 603–609. doi:10.1016/j.copbio.2003.10.011. PMID 14662389.
- Budisa, Nediljko; Pal, Prajna Paramita (1 Jun 2005). "Designing novel spectral classes of proteins with a tryptophan-expanded genetic code". Biol. Chem. 385 (10): 893–904. doi:10.1515/BC.2004.117. PMID 15551863. S2CID 42436705.
- Budisa, Nediljko; Minks, Caroline; Alefelder, Stefan; Wenger, Waltraud; Dong, Fumin; Moroder, Luis; Huber, Huber (1 Jan 1999). "Toward the experimental codon reassignment in vivo: protein building with an expanded amino acid repertoire". FASEB J. 13 (1): 41–51. doi:10.1096/fasebj.13.1.41. PMID 9872928. S2CID 2887572.
- Minks, Caroline; Alefelder, Stefan; Moroder, Luis; Huber, Robert; Budisa, Nediljko (2000). "Towards New Protein Engineering: In Vivo Building and Folding of Protein Shuttles for Drug Delivery and Targeting by the Selective Pressure Incorporation (SPI) Method". Tetrahedron. 56 (48): 9431–9442. doi:10.1016/S0040-4020(00)00827-9.
- Hoesl, M. G.; Oehm, S.; Durkin, P.; Darmon, E.; Peil, L.; Aerni, H.-R.; Rappsilber, J.; Rinehart, J.; Leach, D.; Söll, D.; Budisa, N. (2015). "Chemical evolution of a bacterial proteome". Angewandte Chemie International Edition. 54 (34): 10030–10034. doi:10.1002/anie.201502868. PMC 4782924. PMID 26136259.
- Coyne JA (10 October 1999). "The Truth Is Way Out There". The New York Times. Retrieved 2008-04-06.
- Wu G (May 2009). "Amino acids: metabolism, functions, and nutrition". Amino Acids. 37 (1): 1–17. doi:10.1007/s00726-009-0269-0. PMID 19301095. S2CID 1870305.
External links
- "Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8" - The EMBO Journal
- "Pleiotropic effects of purine auxotrophy inRhizobium meliloti on cell surface molecules" - Springerlink
- "Auxotrophy and Organic Compounds in the Nutrition of Marine Phytoplankton"