BMERB1
bMERB domain containing 1 (or bMERB1) is a gene expressed in humans which has broad expression across the brain. This gene codes for bMERB1 domain-containing protein 1 isoform 1. It is predicted that this gene is involved in actin cytoskeleton regulation, microtubule regulation and glial cell migration.[1]
Gene
Homo sapiens bMERB1 is a protein-coding gene located on Chromosome 16 open reading frame 45 in the cytogenetic band 16p13.11. This gene exists under several aliases, the most common being “MINP” and "c16orf45". bMERB1 spans 2,111 base pairs and contains 6 exons.[2]
mRNA Transcript
| Name | Size (bp) | Protein Length (aa) | Biotype | # of Exons | |
|---|---|---|---|---|---|
| bMERB1-201 | 2111 | 204 | protein coding | 6 | |
| bMERB1-202 | 1865 | 187 | protein coding | 6 | |
| bMERB1-209 | 1854 | 151 | protein coding | 5 | |
| bMERB1-206 | 865 | 175 | protein coding | 5 | |
| bMERB1-203 | 534 | 134 | protein coding | 5 | |
| bMERB1-207 | 497 | 115 | protein coding | 5 | |
| bMERB1-204 | 737 | 88 | nonsense mediated decay | 6 | |
| bMERB1-211 | 622 | 36 | nonsense mediated decay | 5 | |
| bMERB1-208 | 934 | no protein | processed transcript | 4 | |
| bMERB1-205 | 462 | no protein | processed transcript | 2 | |
| bMERB1-210 | 546 | no protein | retained intron | 4 |
Figure 1. This table describes the 11 splice site variants of the bMERB1 transcript.
Protein
Protein Sequence
The bMERB1 isoform 1 protein product is composed of 204 amino acids and it is the longest isoform. Isoform 2 is only 187 amino acids long.[4] In the bMERB1 protein, there are two known regions. One such region (DUF3585) has unknown function and spans around 140 amino acids. Much of the DUF region has been conserved across many species going as far back as birds. A much smaller section of this DUF region was found to be conserved as far back as sharks and rays.[5] The other region is considered a “disordered” region and is only about 20 amino acids long.
Protein Composition
The bMERB1 has a molecular weight of about 24 kilodaltons and has an isoelectronic point of 5.93. There are no positive or negative charge clusters in this sequence. The only repeated amino acid sequence was RLRE, which was only repeated twice. It should also be noted that there are no predicted transmembrane regions in this protein and no hydrophobic regions. bMERB1 can be considered a glycine poor protein.[6]
Gene Level Regulation
Expression Pattern
NCBI human transcriptome analyses reveal that the bMERB1 gene has moderate to high expression in most tissues of the body. However, bMERB1 expression is significantly highest in the brain. More specifically, bMERB1 expression is highest in the amygdala and the frontal cortex. In conclusion, although this gene is not limited to expression in only one tissue, there is a definite pattern of high expression in brain tissue.[1]
Abundance and Localization
The bMERB1 protein has a relatively moderate abundance, and this is true across several other species.[7] In studying antibodies to target bMERB1, it was also determined that the protein is expected to be localized in the cytoplasm.[8]
Transcription Factors
Several transcription factors have been identified to bind to the bMERB1 gene promoter and upstream sequence. It can be noted that a significant number of these transcription factors have known expression patterns in brain tissue. This finding is consistent with the significant bMERB1 gene expression in brain tissue.[9]
Protein Level Regulation
Subcellular Localization
In a PSORTII analysis of BMERB1, it is predicted that this protein is generally located in the nucleus (47.8%), the cytoplasm (26.1%) and the cytoskeleton (13%).[10]
Modifactions
BMERB1 does not obtain many post-translational modifications. A few modifications are predicted with low confidence, most of which being phosphorylation sites.[10] Although the confidence of these predictions is low, similar results were found using PhosphoSitePlus.[11] Using various DTU Health Tech tools, it was also determined that there are no predicted N-glycosylation or myristylation sites.[12] Along with this, PSORTII reported that this protein contains no motifs or transmembrane regions.[10]
Homology and Evolution
| Genus and Species[13] | Common Name | Taxonomic Group | Estimated Divergence (MYA) | Accession Number | Sequence Length (aa) | Sequence Identity (%) | Sequence Similarity (%) | |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | Primates | 0 | NP_149978.1 | 204 | 100 | 100 | |
| Aotus nancymaae | Nancy Ma's Night Monkey | Primates | 43 | XP_012332861.1 | 234 | 86.3 | 86.3 | |
| Ochotona princeps | American Pika | Lagomorpha | 87 | XP_004586852.1 | 204 | 95.1 | 96.6 | |
| Mus Musculus | Mouse | Rodentia | 90 | NP_653101.1 | 203 | 95.1 | 97.1 | |
| Canis lupus familiaris | Dog (beagle) | Carnivora | 94 | XP_038525013 | 204 | 97.06 | 97.1 | |
| Ornithorhynchus anatinus | Duck-Billed Platypus | Monotremata | 180 | XP_001508518.1 | 204 | 86.27 | 92.2 | |
| Nothoprocta perdicaria | Chilean Tinamou | Tinamiformes | 319 | XP_025909891.1 | 204 | 87.7 | 95.1 | |
| Gallus gallus | Chicken | Galliformes | 319 | NP_001264503.1 | 204 | 85.29 | 93.6 | |
| Phaethon lepturus | White-Tailed Seabird | Phaethontiformes | 319 | XP_010280262.1 | 200 | 79.41 | 90.2 | |
| Chelonia mydas | Green Sea Turtle | Testudines | 319 | XP_007058204.1 | 208 | 88 | 93.8 | |
| Varanus komodoensis | Komodo Dragon | Squamata | 319 | XP_044289683.1 | 207 | 79.23 | 90.8 | |
| Eleutherodactylus coqui | Puerto Rican Coqui | Anura | 353 | XP_018419715 | 210 | 72.9 | 84.8 | |
| Stegastes partitus | Perch-Like Fish | Perciformes | 464 | XP_008278140.1 | 200 | 68.6 | 81.6 | |
| Chiloscyllium plagiosum | Carpet Shark | Orectolobiformes | 464 | XP_043568064.1 | 206 | 64.08 | 78.6 | |
| Petromyzon marinus | Sea Lamprey | Sea lamprey | 615 | XP_032832012.1 | 208 | 46.1 | 65.3 | |
| Danio rerio | Zebrafish | Actinoptergyii (bony fish) | 435 | NP_001028910.1 | 184 | 54.9 | 71.1 |
Paralogs
There are no paralogs of bMERB1.[13]
Mutation Rate of bMERB1
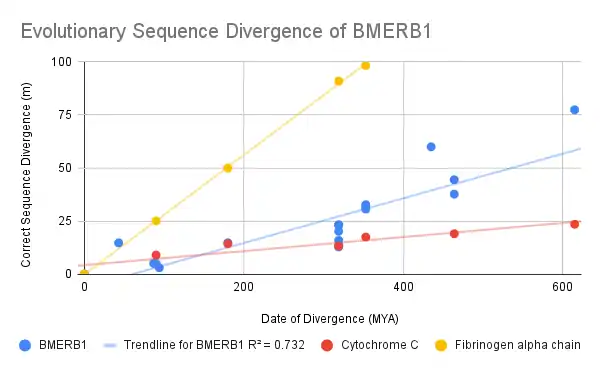
From this graph, it can be noted that the rate of sequence divergence in BMERB1 is between the fast-diverging protein Fibrinogen alpha chain and the slow-diverging Cytochrome C. This would indicate that BMERB1 is not under evolutionary pressure to mutate at an accelerated rate but is also not being prevented from mutating.
Interacting Proteins
Several bMERB1 protein interactions have been identified and their functions also discovered. Generally, the bMERB1 protein interactions have functions likely to be involved in actin cytoskeleton maintenance and microtubule formation/maintenance.[14]
Clinical Significance
No pathogenic genetic mutations have been identified in the bMERB1 sequence.[15]
References
- "BMERB1 bMERB domain containing 1 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2022-07-29.
- "Homo sapiens bMERB domain containing 1 (BMERB1), transcript variant 1, mRNA". 2021-06-26.
- "BMERB1 Gene - GeneCards | MERB1 Protein | MERB1 Antibody". www.genecards.org. Retrieved 2022-07-29.
- "UniProt". www.uniprot.org. Retrieved 2022-07-29.
- "EMBOSS Needle < Pairwise Sequence Alignment < EMBL-EBI". www.ebi.ac.uk. Retrieved 2022-07-29.
- "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2022-07-29.
- "PAXdb: Protein Abundance Database". www.pax-db.org. Retrieved 2022-07-29.
- "CP045 Antibody (PA5-67599)". www.thermofisher.com. Retrieved 2022-07-29.
- "The Human Protein Atlas". www.proteinatlas.org. Retrieved 2022-07-29.
- "PSORT II Prediction". psort.hgc.jp. Retrieved 2022-07-29.
- "PhosphoSitePlus". www.phosphosite.org. Retrieved 2022-07-29.
- "Department of Health Technology - DTU Health Tech". www.healthtech.dtu.dk. Retrieved 2022-07-29.
- "Nucleotide BLAST: Search nucleotide databases using a nucleotide query". blast.ncbi.nlm.nih.gov. Retrieved 2022-07-29.
- "14 items (human) - STRING interaction network". string-db.org. Retrieved 2022-07-29.
- "Variation Viewer". www.ncbi.nlm.nih.gov. Retrieved 2022-07-29.