BPIFA3
BPI fold containing family A, member 3 (BPIFA3) is a protein that in humans is encoded by the BPIFA3 gene.[5] The gene is also known as SPLUNC3 and C20orf71 in humans and the orthologous gene in mice is 1700058C13Rik.[6] There are multiple variants of the BPIFA3 projected to be a secreted protein.[7] It is very highly expressed in testis with little or no expression in other tissues. The Human Protein Atlas project[8] and Mouse ENCODE Consortium[9] report RNA-Seq expression at RPKM levels (reads per kilobases of transcript per 1 million mapped reads ) of 29.1 for human testis and 69.4 for mouse, but 0 for all other tissues.[5][6] Similarly, the Bgee consortium,[10] using multiple techniques in addition to RNA-Seq, reports a relative Expression Score of 95.8 out of 100 for testis and 99.0 for sperm in humans; however low levels of BPIFA3 between 20 and 30 were seen for a variety of tissues such as muscle, glands, prostate, nervous system, and skin.[11]
| BPIFA3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | BPIFA3, C20orf71, SPLUNC3, BPI fold containing family A member 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1920638 HomoloGene: 52268 GeneCards: BPIFA3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Superfamily
BPIFA3 is a member of a BPI fold protein superfamily defined by the presence of the bactericidal/permeability-increasing protein fold (BPI fold) which is formed by two similar domains in a "boomerang" shape.[12] This superfamily is also known as the BPI/LBP/PLUNC family or the BPI/LPB/CETP family.[13] The BPI fold creates apolar binding pockets that can interact with hydrophobic and amphipathic molecules, such as the acyl carbon chains of lipopolysaccharide found on Gram-negative bacteria, but members of this family may have many other functions.
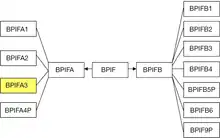
Genes for the BPI/LBP/PLUNC superfamily are found in all vertebrate species, including distant homologs in non-vertebrate species such as insects, mollusks, and roundworms.[14][15] Within that broad grouping is the BPIF gene family whose members encode the BPI fold structural motif and are found clustered on a single chromosome, e.g., Chromosome 20 in humans, Chromosome 2 in mouse, Chromosome 3 in rat, Chromosome 17 in pig, Chromosome 13 in cow. The BPIF gene family is split into two groupings, BPIFA and BPIFB. In humans, BIPFA consists of 3 protein encoding genes BPIFA1, BPIFA2, BPIFA3, and 1 pseudogene BPIFA4P; while BPIFB consists of 5 protein encoding genes BPIFB1, BPIFB2, BPIFB3, BPIFB4, BPIFB6 and 2 pseudogenes BPIFB5P, BPIFB9P. What appears as pseudogenes in humans may appear as fully functional genes in other species.
In humans, the BPIFA3 gene was first identified as a human PLUNC-related gene[16] and later in mouse as 1700058C13Rik.[17] Both were recognized as BPIF gene family members and were renamed BPIFA3 and Bpifa3, respectively.
Function
Little is known about BPIFA3's function. Due to its high degree of similarity to other members of the BPI/LBP/PLUNC superfamily, it is predicted to have binding activity to lipids such as phospholipids and lipopolysaccharides but this has never been confirmed.[18]
A putative function of BPIFA3 in bacterial host defense has been suggested. Despite a near-exclusive expression of BPIFA3 in testis, only a single report implicates it in any normal or pathological states and that involves Otitis media (OM) in the ear.[19] OM is a common disease of early childhood characterized by inflammation of the middle ear cavity and an effort was made to identify genetic risk factors. A genome-wide association study was done in the "Raine Study," a longitudinal birth cohort of 2,868 children in which 1532 blood samples drawn at 1,2, and 3 years of age were analyzed for gene expression correlated to a susceptibility to OM. Unexpectedly, BPIFA3 and BPIFA1 were the top hits, along with CAPN14 and GALNT14 genes, none of which has previously been implicated in OM. The investigators noted that BPIF gene family members encode proteins involved in the early recognition of bacterial pathogens as part of the host defense at the nasopharyngeal, oral and lung entrances, and that expression of BPIFA3 and BPIFA1 may be a logical consequence of chronic bacterial presence in the pus filled, OM infected ear.
Model organisms
| Characteristic | Phenotype |
|---|---|
| Homozygote viability | Normal |
| Fertility | Normal |
| Body weight | Normal |
| Anxiety | Normal |
| Neurological assessment | Normal |
| Grip strength | Normal |
| Hot plate | Normal |
| Dysmorphology | Normal |
| Indirect calorimetry | Normal |
| Glucose tolerance test | Normal |
| Auditory brainstem response | Normal |
| DEXA | Normal |
| Radiography | Normal |
| Body temperature | Normal |
| Eye morphology | Normal |
| Clinical chemistry | Normal |
| Haematology | Normal |
| Micronucleus test | Normal |
| Heart weight | Normal |
| Skin Histopathology | Normal |
| Eye Histopathology | Normal |
| Salmonella infection | Normal[20] |
| Citrobacter infection | Normal[21] |
| All tests and analysis from[22][23] | |
Model organisms have been used in the study of BPIFA3 function. A conditional knockout mouse line, called 1700058C13Riktm1a(KOMP)Wtsi[24][25] was generated as part of the International Knockout Mouse Consortium program — a high-throughput mutagenesis project to generate and distribute animal models of disease to interested scientists — at the Wellcome Trust Sanger Institute.[26][27][28]
Male and female animals underwent a standardized phenotypic screen to determine the effects of deletion.[22][29] Twenty three tests were carried out on mutant mice, but no significant abnormalities were observed.[22]
References
- GRCh38: Ensembl release 89: ENSG00000131059 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000027482 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "BPIFA3 BPI fold containing family A member 3 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 17 February 2023.
- "Bpifa3 BPI fold containing family A, member 3 [Mus musculus (house mouse)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 17 February 2023.
- "Q9BQP9 - BPIA3_HUMAN". www.uniprot.org. Retrieved 17 February 2023.
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. (February 2014). "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics". Molecular & Cellular Proteomics. 13 (2): 397–406. doi:10.1074/mcp.M113.035600. PMC 3916642. PMID 24309898.
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. (November 2014). "A comparative encyclopedia of DNA elements in the mouse genome". Nature. 515 (7527): 355–64. Bibcode:2014Natur.515..355.. doi:10.1038/nature13992. PMC 4266106. PMID 25409824.
- Bastian FB, Roux J, Niknejad A, Comte A, Fonseca Costa SS, de Farias TM, et al. (January 2021). "The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals". Nucleic Acids Research. 49 (D1): D831–D847. doi:10.1093/nar/gkaa793. PMC 7778977. PMID 33037820.
- "Gene: BPIFA3 - ENSG00000131059". bgee.org. The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals. Retrieved 17 February 2023.
- Beamer LJ, Carroll SF, Eisenberg D (April 1998). "The BPI/LBP family of proteins: a structural analysis of conserved regions". Protein Science. 7 (4): 906–914. doi:10.1002/pro.5560070408. PMC 2143972. PMID 9568897.
- "CDD Conserved Protein Domain Family: BPI". www.ncbi.nlm.nih.gov.
- Beamer LJ, Fischer D, Eisenberg D (July 1998). "Detecting distant relatives of mammalian LPS-binding and lipid transport proteins". Protein Science. 7 (7): 1643–1646. doi:10.1002/pro.5560070721. PMC 2144061. PMID 9684900.
- Bingle CD, Seal RL, Craven CJ (August 2011). "Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily". Biochemical Society Transactions. 39 (4): 977–983. doi:10.1042/BST0390977. PMC 3196848. PMID 21787333.
- Bingle, CD; Craven, CJ (15 April 2002). "PLUNC: A novel family of candidate host defense proteins expressed in the upper airways and nasopharynx". Human Molecular Genetics. 11 (8): 937–943. doi:10.1093/hmg/11.8.937. PMID 11971875.
- Oakley MS, McCutchan TF, Anantharaman V, Ward JM, Faucette L, Erexson C, et al. (October 2008). "Host biomarkers and biological pathways that are associated with the expression of experimental cerebral malaria in mice". Infection and Immunity. 76 (10): 4518–29. doi:10.1128/IAI.00525-08. PMC 2546852. PMID 18644885.
- "BPIFA3 Gene - GeneCards". www.genecards.org. Retrieved 17 February 2023.
- Rye MS, Warrington NM, Scaman ES, Vijayasekaran S, Coates HL, Anderson D, Pennell CE, Blackwell JM, Jamieson SE (2012). "Genome-wide association study to identify the genetic determinants of otitis media susceptibility in childhood". PLOS ONE. 7 (10): e48215. Bibcode:2012PLoSO...748215R. doi:10.1371/journal.pone.0048215. PMC 3485007. PMID 23133572.
- "Salmonella infection data for 1700058C13Rik". Wellcome Trust Sanger Institute.
- "Citrobacter infection data for 1700058C13Rik". Wellcome Trust Sanger Institute.
- Gerdin AK (2010). "The Sanger Mouse Genetics Programme: High throughput characterisation of knockout mice". Acta Ophthalmologica. 88 (S248). doi:10.1111/j.1755-3768.2010.4142.x. S2CID 85911512.
- Mouse Resources Portal, Wellcome Trust Sanger Institute.
- "International Knockout Mouse Consortium".
- "Mouse Genome Informatics".
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. (June 2011). "A conditional knockout resource for the genome-wide study of mouse gene function". Nature. 474 (7351): 337–342. doi:10.1038/nature10163. PMC 3572410. PMID 21677750.
- Dolgin E (June 2011). "Mouse library set to be knockout". Nature. 474 (7351): 262–263. doi:10.1038/474262a. PMID 21677718.
- Collins FS, Rossant J, Wurst W (January 2007). "A mouse for all reasons". Cell. 128 (1): 9–13. doi:10.1016/j.cell.2006.12.018. PMID 17218247. S2CID 18872015.
- van der Weyden L, White JK, Adams DJ, Logan DW (June 2011). "The mouse genetics toolkit: revealing function and mechanism". Genome Biology. 12 (6): 224. doi:10.1186/gb-2011-12-6-224. PMC 3218837. PMID 21722353.
External links
- Human BPIFA3 genome location and BPIFA3 gene details page in the UCSC Genome Browser.
- BPIFA3+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)