Neurogenetics
Neurogenetics studies the role of genetics in the development and function of the nervous system. It considers neural characteristics as phenotypes (i.e. manifestations, measurable or not, of the genetic make-up of an individual), and is mainly based on the observation that the nervous systems of individuals, even of those belonging to the same species, may not be identical. As the name implies, it draws aspects from both the studies of neuroscience and genetics, focusing in particular how the genetic code an organism carries affects its expressed traits. Mutations in this genetic sequence can have a wide range of effects on the quality of life of the individual. Neurological diseases, behavior and personality are all studied in the context of neurogenetics. The field of neurogenetics emerged in the mid to late 20th century with advances closely following advancements made in available technology. Currently, neurogenetics is the center of much research utilizing cutting edge techniques.
History
The field of neurogenetics emerged from advances made in molecular biology, genetics and a desire to understand the link between genes, behavior, the brain, and neurological disorders and diseases. The field started to expand in the 1960s through the research of Seymour Benzer, considered by some to be the father of neurogenetics.[1]

His pioneering work with Drosophila helped to elucidate the link between circadian rhythms and genes, which led to further investigations into other behavior traits. He also started conducting research in neurodegeneration in fruit flies in an attempt to discover ways to suppress neurological diseases in humans. Many of the techniques he used and conclusions he drew would drive the field forward.[2]
Early analysis relied on statistical interpretation through processes such as LOD (logarithm of odds) scores of pedigrees and other observational methods such as affected sib-pairs, which looks at phenotype and IBD (identity by descent) configuration. Many of the disorders studied early on including Alzheimer's, Huntington's and amyotrophic lateral sclerosis (ALS) are still at the center of much research to this day.[3] By the late 1980s new advances in genetics such as recombinant DNA technology and reverse genetics allowed for the broader use of DNA polymorphisms to test for linkage between DNA and gene defects. This process is referred to sometimes as linkage analysis.[4][5] By the 1990s ever advancing technology had made genetic analysis more feasible and available. This decade saw a marked increase in identifying the specific role genes played in relation to neurological disorders. Advancements were made in but not limited to: Fragile X syndrome, Alzheimer's, Parkinson's, epilepsy and ALS.[6]
Neurological disorders
While the genetic basis of simple diseases and disorders has been accurately pinpointed, the genetics behind more complex, neurological disorders is still a source of ongoing research. New developments such as the genome wide association studies (GWAS) have brought vast new resources within grasp. With this new information genetic variability within the human population and possibly linked diseases can be more readily discerned.[7] Neurodegenerative diseases are a more common subset of neurological disorders, with examples being Alzheimer's disease and Parkinson's disease. Currently no viable treatments exist that actually reverse the progression of neurodegenerative diseases; however, neurogenetics is emerging as one field that might yield a causative connection. The discovery of linkages could then lead to therapeutic drugs, which could reverse brain degeneration.[8]
Gene sequencing
One of the most noticeable results of further research into neurogenetics is a greater knowledge of gene loci that show linkage to neurological diseases. The table below represents a sampling of specific gene locations identified to play a role in selected neurological diseases based on prevalence in the United States.[9][10][11][12]
| Gene loci | Neurological disease |
|---|---|
| APOE ε4, PICALM[10] | Alzheimer's disease |
| DR15, DQ6[11] | Multiple sclerosis |
| LRRK2, PARK2, PARK7[9] | Parkinson's disease |
| HTT[12] | Huntington's disease |
Methods of research
Statistical analysis
Logarithm of odds (LOD) is a statistical technique used to estimate the probability of gene linkage between traits. LOD is often used in conjunction with pedigrees, maps of a family's genetic make-up, in order to yield more accurate estimations. A key benefit of this technique is its ability to give reliable results in both large and small sample sizes, which is a marked advantage in laboratory research.[13][14]
Quantitative trait loci (QTL) mapping is another statistical method used to determine the chromosomal positions of a set of genes responsible for a given trait. By identifying specific genetic markers for the genes of interest in a recombinant inbred strain, the amount of interaction between these genes and their relation to the observed phenotype can be determined through complex statistical analysis. In a neurogenetics laboratory, the phenotype of a model organisms is observed by assessing the morphology of their brain through thin slices.[15] QTL mapping can also be carried out in humans, though brain morphologies are examined using nuclear magnetic resonance imaging (MRI) rather than brain slices. Human beings pose a greater challenge for QTL analysis because the genetic population cannot be as carefully controlled as that of an inbred recombinant population, which can result in sources of statistical error.[16]
Recombinant DNA
Recombinant DNA is an important method of research in many fields, including neurogenetics. It is used to make alterations to an organism's genome, usually causing it to over- or under-express a certain gene of interest, or express a mutated form of it. The results of these experiments can provide information on that gene's role in the organism's body, and it importance in survival and fitness. The hosts are then screened with the aid of a toxic drug that the selectable marker is resistant to. The use of recombinant DNA is an example of a reverse genetics, where researchers create a mutant genotype and analyze the resulting phenotype. In forward genetics, an organism with a particular phenotype is identified first, and its genotype is then analyzed.[17][18]
Animal research


Model organisms are an important tool in many areas of research, including the field of neurogenetics. By studying creatures with simpler nervous systems and with smaller genomes, scientists can better understand their biological processes and apply them to more complex organisms, such as humans. Due to their low-maintenance and highly mapped genomes, mice, Drosophila,[19] and C. elegans[20] are very common. Zebrafish[21] and prairie voles[22] have also become more common, especially in the social and behavioral scopes of neurogenetics.
In addition to examining how genetic mutations affect the actual structure of the brain, researchers in neurogenetics also examine how these mutations affect cognition and behavior. One method of examining this involves purposely engineering model organisms with mutations of certain genes of interest. These animals are then classically conditioned to perform certain types of tasks, such as pulling a lever in order to gain a reward. The speed of their learning, the retention of the learned behavior, and other factors are then compared to the results of healthy organisms to determine what kind of an effect – if any – the mutation has had on these higher processes. The results of this research can help identify genes that may be associated with conditions involving cognitive and learning deficiencies.[23]
Human research
Many research facilities seek out volunteers with certain conditions or illnesses to participate in studies. Model organisms, while important, cannot completely model the complexity of the human body, making volunteers a key part to the progression of research. Along with gathering some basic information about medical history and the extent of their symptoms, samples are taken from the participants, including blood, cerebrospinal fluid, and/or muscle tissue. These tissue samples are then genetically sequenced, and the genomes are added to current database collections. The growth of these data bases will eventually allow researchers to better understand the genetic nuances of these conditions and bring therapy treatments closer to reality. Current areas of interest in this field have a wide range, spanning anywhere from the maintenance of circadian rhythms, the progression of neurodegenerative disorders, the persistence of periodic disorders, and the effects of mitochondrial decay on metabolism.[24]
Genome-wide association studies
Such databases are used in genome-wide association studies (GWAS). Examples of phenotypes investigated by notable neurogenetics GWAS include:
- Language-related skills: a meta-analysis of GWAS reported genetic factors of, the so far uniquely human, language-related capabilities, in particular factors of differences in skill-levels of five tested traits. It also used neuroanatomy/neuroimaging data.[25][26]
Behavioral neurogenetics
Advances in molecular biology techniques and the species-wide genome project have made it possible to map out an individual's entire genome. Whether genetic or environmental factors are primarily responsible for an individual's personality has long been a topic of debate.[27][28] Thanks to the advances being made in the field of neurogenetics, researchers have begun to tackle this question by beginning to map out genes and correlate them to different personality traits.[27] There is little to no evidence to suggest that the presence of a single gene indicates that an individual will express one style of behavior over another; rather, having a specific gene could make one more predisposed to displaying this type of behavior. It is starting to become clear that most genetically influenced behaviors are due to the effects of many variants within many genes, in addition to other neurological regulating factors like neurotransmitter levels. Due to fact that many behavioral characteristics have been conserved across species for generations, researchers are able to use animal subjects such as mice and rats, but also fruit flies, worms, and zebrafish,[19][20] to try to determine specific genes that correlate to behavior and attempt to match these with human genes.[29]
Cross-species gene conservation
While it is true that variation between species can appear to be pronounced, at their most basic they share many similar behavior traits which are necessary for survival. Such traits include mating, aggression, foraging, social behavior and sleep patterns. This conservation of behavior across species has led biologists to hypothesize that these traits could possibly have similar, if not the same, genetic causes and pathways. Studies conducted on the genomes of a plethora of organisms have revealed that many organisms have homologous genes, meaning that some genetic material has been conserved between species. If these organisms shared a common evolutionary ancestor, then this might imply that aspects of behavior can be inherited from previous generations, lending support to the genetic causes – as opposed to the environmental causes – of behavior.[28] Variations in personalities and behavioral traits seen amongst individuals of the same species could be explained by differing levels of expression of these genes and their corresponding proteins.[29]
Aggression
There is also research being conducted on how an individual's genes can cause varying levels of aggression and aggression control .

Throughout the animal kingdom, varying styles, types and levels of aggression can be observed leading scientists to believe that there might be a genetic contribution that has conserved this particular behavioral trait.[30] For some species varying levels of aggression have indeed exhibited direct correlation to a higher level of Darwinian fitness.[31]
Development
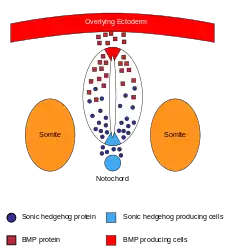
A great deal of research has been done on the effects of genes and the formation of the brain and the central nervous system. The following wiki links may prove helpful:
There are many genes and proteins that contribute to the formation and development of the central nervous system, many of which can be found in the aforementioned links. Of particular importance are those that code for BMPs, BMP inhibitors and SHH. When expressed during early development, BMP's are responsible for the differentiation of epidermal cells from the ventral ectoderm. Inhibitors of BMPs, such as NOG and CHRD, promote differentiation of ectoderm cells into prospective neural tissue on the dorsal side. If any of these genes are improperly regulated, then proper formation and differentiation will not occur. BMP also plays a very important role in the patterning that occurs after the formation of the neural tube. Due to the graded response the cells of the neural tube have to BMP and Shh signaling, these pathways are in competition to determine the fate of preneural cells. BMP promotes dorsal differentiation of pre-neural cells into sensory neurons and Shh promotes ventral differentiation into motor neurons. There are many other genes that help to determine neural fate and proper development include, RELN, SOX9, WNT, Notch and Delta coding genes, HOX, and various cadherin coding genes like CDH1 and CDH2.[32]
Some recent research has shown that the level of gene expression changes drastically in the brain at different periods throughout the life cycle. For example, during prenatal development the amount of mRNA in the brain (an indicator of gene expression) is exceptionally high, and drops to a significantly lower level not long after birth. The only other point of the life cycle during which expression is this high is during the mid- to late-life period, during 50–70 years of age. While the increased expression during the prenatal period can be explained by the rapid growth and formation of the brain tissue, the reason behind the surge of late-life expression remains a topic of ongoing research.[33]
Current research
Neurogenetics is a field that is rapidly expanding and growing. The current areas of research are very diverse in their focuses. One area deals with molecular processes and the function of certain proteins, often in conjunction with cell signaling and neurotransmitter release, cell development and repair, or neuronal plasticity. Behavioral and cognitive areas of research continue to expand in an effort to pinpoint contributing genetic factors. As a result of the expanding neurogenetics field a better understanding of specific neurological disorders and phenotypes has arisen with direct correlation to genetic mutations. With severe disorders such as epilepsy, brain malformations, or mental retardation a single gene or causative condition has been identified 60% of the time; however, the milder the intellectual handicap the lower chance a specific genetic cause has been pinpointed. Autism for example is only linked to a specific, mutated gene about 15–20% of the time while the mildest forms of mental handicaps are only being accounted for genetically less than 5% of the time. Research in neurogenetics has yielded some promising results, though, in that mutations at specific gene loci have been linked to harmful phenotypes and their resulting disorders. For instance a frameshift mutation or a missense mutation at the DCX gene location causes a neuronal migration defect also known as lissencephaly. Another example is the ROBO3 gene where a mutation alters axon length negatively impacting neuronal connections. Horizontal gaze palsy with progressive scoliosis (HGPPS) accompanies a mutation here.[34] These are just a few examples of what current research in the field of neurogenetics has achieved.[35]
See also
- Journals
References
- "Olympians of Science: A Display of Medals and Awards". California Institute of Technology. Retrieved 5 December 2011.
- "Neurogenetics Pioneer Seymour Benzer Dies". California Institute of Technology. Archived from the original on 20 January 2012. Retrieved 5 December 2011.
- Gershon ES, Goldin LR (1987). "The outlook for linkage research in psychiatric disorders". Journal of Psychiatric Research. 21 (4): 541–50. doi:10.1016/0022-3956(87)90103-8. PMID 3326940.
- Tanzi RE (October 1991). "Genetic linkage studies of human neurodegenerative disorders". Current Opinion in Neurobiology. 1 (3): 455–61. doi:10.1016/0959-4388(91)90069-J. PMID 1840379. S2CID 19732183.
- Greenstein P, Bird TD (September 1994). "Neurogenetics. Triumphs and challenges". The Western Journal of Medicine. 161 (3): 242–5. PMC 1011404. PMID 7975561.
- Tandon PN (September 2000). "The decade of the brain: a brief review". Neurology India. 48 (3): 199–207. PMID 11025621.
- Simón-Sánchez J, Singleton A (November 2008). "Genome-wide association studies in neurological disorders". The Lancet. Neurology. 7 (11): 1067–72. doi:10.1016/S1474-4422(08)70241-2. PMC 2824165. PMID 18940696.
- Kumar A, Cookson MR (June 2011). "Role of LRRK2 kinase dysfunction in Parkinson disease". Expert Reviews in Molecular Medicine. 13 (20): e20. doi:10.1017/S146239941100192X. PMC 4672634. PMID 21676337.
- "Parkinson disease". NIH. Retrieved 6 December 2011.
- "Alzheimer's Disease Genetics Fact Sheet". NIH. Archived from the original on 28 November 2011. Retrieved 6 December 2011.
- "Multiple Sclerosis". NIH.
- "Huntington Disease". Genetics Home Reference. NIH. 15 April 2020.
- Morton NE (April 1996). "Logarithm of odds (lods) for linkage in complex inheritance". Proceedings of the National Academy of Sciences of the United States of America. 93 (8): 3471–6. Bibcode:1996PNAS...93.3471M. doi:10.1073/pnas.93.8.3471. PMC 39633. PMID 8622960.
- Helms T (2000). "Logarithm of Odds in Advanced Genetics". North Dakota State University. Archived from the original on 26 January 2006.
- R. W. Williams (1998) Neuroscience Meets Quantitative Genetics: Using Morphometric Data to Map Genes that Modulate CNS Architecture.
- Bartley AJ, Jones DW, Weinberger DR (February 1997). "Genetic variability of human brain size and cortical gyral patterns". Brain. 120 ( Pt 2) (2): 257–69. doi:10.1093/brain/120.2.257. PMID 9117373.
- Kuure-Kinsey M, McCooey B (Fall 2000). "The Basics of Recombinant DNA". RPI.edu.
- Ambrose, Victor (2011). Reverse Genetics.
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, et al. (July 2008). "Tools for neuroanatomy and neurogenetics in Drosophila". Proceedings of the National Academy of Sciences of the United States of America. 105 (28): 9715–20. Bibcode:2008PNAS..105.9715P. doi:10.1073/pnas.0803697105. PMC 2447866. PMID 18621688.
- Rand JB, Duerr JS, Frisby DL (December 2000). "Neurogenetics of vesicular transporters in C. elegans". FASEB Journal. 14 (15): 2414–22. doi:10.1096/fj.00-0313rev. PMID 11099459. S2CID 17074233.
- Burgess HA, Granato M (November 2008). "The neurogenetic frontier--lessons from misbehaving zebrafish". Briefings in Functional Genomics & Proteomics. 7 (6): 474–82. doi:10.1093/bfgp/eln039. PMC 2722256. PMID 18836206.
- McGraw LA, Young LJ (February 2010). "The prairie vole: an emerging model organism for understanding the social brain". Trends in Neurosciences. 33 (2): 103–9. doi:10.1016/j.tins.2009.11.006. PMC 2822034. PMID 20005580.
- Neurogenetics and Behavior Center. Johns Hopkins U, 2011. Web. 29 Oct. 2011.
- Fu YH, Ptacek L (29 October 2011). "Research Projects". Fu and Ptacek's Laboratories of Neurogenetics. U of California, San Francisco.
- "Massive genome study informs the biology of reading and language". Max Planck Society via medicalxpress.com. Retrieved 18 September 2022.
- Eising, Else; Mirza-Schreiber, Nazanin; de Zeeuw, Eveline L.; Wang, Carol A.; Truong, Dongnhu T.; Allegrini, Andrea G.; Shapland, Chin Yang; Zhu, Gu; Wigg, Karen G.; Gerritse, Margot L.; et al. (30 August 2022). "Genome-wide analyses of individual differences in quantitatively assessed reading- and language-related skills in up to 34,000 people". Proceedings of the National Academy of Sciences. 119 (35): e2202764119. Bibcode:2022PNAS..11902764E. doi:10.1073/pnas.2202764119. ISSN 0027-8424. PMC 9436320. PMID 35998220.
- Congdon E, Canli T (December 2008). "A neurogenetic approach to impulsivity". Journal of Personality (Print). 76 (6): 1447–84. doi:10.1111/j.1467-6494.2008.00528.x. PMC 2913861. PMID 19012655.
- Kimura M, Higuchi S (April 2011). "Genetics of alcohol dependence". Psychiatry and Clinical Neurosciences (Print). 65 (3): 213–25. doi:10.1111/j.1440-1819.2011.02190.x. PMID 21507127.
- Reaume CJ, Sokolowski MB (July 2011). "Conservation of gene function in behaviour". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 366 (1574): 2100–10. doi:10.1098/rstb.2011.0028. PMC 3130371. PMID 21690128.
- Zwarts L, Magwire MM, Carbone MA, Versteven M, Herteleer L, Anholt RR, et al. (October 2011). "Complex genetic architecture of Drosophila aggressive behavior". Proceedings of the National Academy of Sciences of the United States of America. 108 (41): 17070–5. Bibcode:2011PNAS..10817070Z. doi:10.1073/pnas.1113877108. PMC 3193212. PMID 21949384.
- Oliveira RF, Silva JF, Simões JM (June 2011). "Fighting zebrafish: characterization of aggressive behavior and winner-loser effects". Zebrafish (Print). 8 (2): 73–81. doi:10.1089/zeb.2011.0690. PMID 21612540.
- Alberts; et al. (2008). Molecular Biology of the Cell (5th ed.). Garland Science. pp. 1139–1480. ISBN 978-0-8153-4105-5.
- Sanders L (2011). "Brain gene activity changes through life".
- Walsh CA, Engle EC (October 2010). "Allelic diversity in human developmental neurogenetics: insights into biology and disease". Neuron. 68 (2): 245–53. doi:10.1016/j.neuron.2010.09.042. PMC 3010396. PMID 20955932.
- "This Week In the Journal." The Journal of Neuroscience.

