Blasticidin S
Blasticidin S is an antibiotic that is used in biology research for selecting cells in cell culture. Cells of interest can express the blasticidin resistance genes BSD or bsr, and can then survive treatment with the antibiotic. Blasticidin S is a nucleoside analogue antibiotic, resembling the nucleoside cytidine. Blasticidin works against human cells, fungi, and bacteria, all by disrupting protein translation. It was originally described by Japanese researchers in the 1950s seeking antibiotics for rice blast fungus.
 | |
| Names | |
|---|---|
| Preferred IUPAC name
(2S,3S,6R)-3-{(3S)-3-Amino-5-[carbamimidoyl(methyl)amino]pentanamido}-6-(4-amino-2-oxopyrimidin-1(2H)-yl)-3,6-dihydro-2H-pyran-2-carboxylic acid | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.109.057 |
| KEGG | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C17H26N8O5 | |
| Molar mass | 422.44 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Chemistry
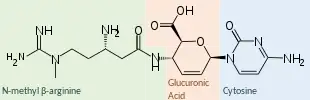
A nucleoside analog, blasticidin S resembles the nucleoside cytidine. The chemical structure consists of a cytosine molecule, linked to a glucuronic acid-derived ring, linked in turn to the peptide N-methyl β-arginine.[1]
Uses
Blasticidin S is widely used in cell culture for selecting and maintaining genetically manipulated cells. Cells of interest express the blasticidin S resistance genes BSD or bsr, and can then survive blasticidin S being added to the culture media.[2] Blasticidin S is typically used at 2–300 micrograms per milliliter of media, depending on the type of cell being grown.[2]
Mechanism of action
Blasticidin prevents the growth of both eukaryotic and prokaryotic cells. It works by inhibiting termination step of translation and peptide bond formation (to lesser extent) by the ribosome. This means that cells can no longer produce new proteins through translation of mRNA. It is competitive with puromycin suggesting a highly similar binding site.[3]
Biosynthesis
The first step in blasticidin S biosynthesis is the combination of UDP-glucuronic acid with cytosine to form cytosylglucuronic acid (CGA). Given the product name, the enzyme that performs this combination is called CGA synthase.[1]
Cosmid cloning experiments from the Blasticidin S producer Streptomyces griseochromogenes, followed by evaluation of the putative biosynthetic gene cluster via heterologous reconstitution of Blasticidin S production in Streptomyces lividans, indicated that a 20 Kbp gene cluster with 19 genes, plus possibly a peptidase outside the gene cluster that acts on the final leucylblasticidin S (LBS) intermediate, was sufficient for reconstitution of Blasticidin S biosynthesis.[3]
Resistance genes
Resistance to blasticidin S can be conferred by either of two deaminases: BSD, originally isolated from Aspergillus terreus or bsr, isolated from Bacillus cereus. Both deaminases work by modifying blasticidin S directly, replacing the amine on the cytosine ring with a hydroxyl group, resulting in the inactive deaminohydroxy-blasticin S.[2][4]
bsr and BSD are the most commonly used resistance genes. The proteins produced from these genes enable the cells carrying them to produce proteins in the presence of blasticidin.
History
In the 1950s, a drug screening program was designed in Japan to discover a new antibiotic that prevents blast disease by the fungus Magnaporthe grisea.[5]
References
- Niu G, Tan H (February 2015). "Nucleoside antibiotics: biosynthesis, regulation, and biotechnology". Trends Microbiol. 23 (2): 110–9. doi:10.1016/j.tim.2014.10.007. PMID 25468791.
- "Blasticidin S HCl Protocols". ThermoFisher Scientific. Retrieved 22 April 2021.
- Cone, Martha C.; Yin, Xihou; Grochowski, Laura L.; Parker, Morgan R.; Zabriskie, T. Mark (2003-09-04). "The Blasticidin S Biosynthesis Gene Cluster from Streptomyces griseochromogenes: Sequence Analysis, Organization, and Initial Characterization". ChemBioChem. Wiley. 4 (9): 821–828. doi:10.1002/cbic.200300583. ISSN 1439-4227.
- Kimura M, Yamamoto M, Furuichi M, Kumasaka T, Yamaguchi I (2002). "An unexpected gift from fungicide metabolism studies: blasticidin S deaminase (BSD) from Aspergillus terreus". Progress in Biotechnology. 22: 55–60. doi:10.1016/S0921-0423(02)80043-0.
- Natural Products Isolation: Separation Methods for Antimicrobials, Antivirals, and Enzyme Inhibitors. Wagman G. H., Elsevier R. C.; p. 191 (1988).