Branch migration
Branch migration is the process by which base pairs on homologous DNA strands are consecutively exchanged at a Holliday junction, moving the branch point up or down the DNA sequence.[1] Branch migration is the second step of genetic recombination, following the exchange of two single strands of DNA between two homologous chromosomes.[2] The process is random, and the branch point can be displaced in either direction on the strand, influencing the degree of which the genetic material is exchanged.[1] Branch migration can also be seen in DNA repair and replication, when filling in gaps in the sequence. It can also be seen when a foreign piece of DNA invades the strand.[2]
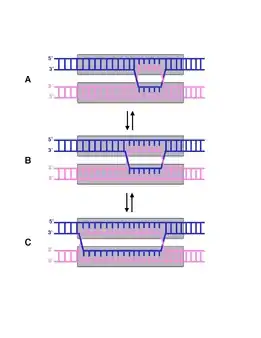
Mechanism
The mechanism for branch migration differs between prokaryotes and eukaryotes.[2]
Prokaryotes

The mechanism for prokaryotic branch migration has been studied many times in Escherichia coli.[2] In E. coli, the proteins RuvA and RuvB come together and form a complex that facilitates the process in a number of ways. RuvA is a tetramer and binds to the DNA at the Holliday junction when it is in the open X form. The protein binds in a way that the DNA entering/departing the junction is still free to rotate and slide through. RuvA has a domain with acidic amino acid residues that interfere with the base pairs in the centre of the junction. This forces the base pairs apart so that they can re-anneal with base pairs on the homologous strands.[3]
In order for migration to occur, RuvA must be associated with RuvB and ATP. RuvB has the ability to hydrolyze ATP, driving the movement of the branch point. RuvB is a hexamer with helicase activity, and also binds the DNA. As ATP is hydrolyzed, RuvB rotates the recombined strands while pulling them out of the junction, but does not separate the strands as helicase would.[3]
The final step in branch migration is called resolution and requires the protein RuvC. The protein is a dimer, and will bind to the Holliday junction when it takes on the stacked X form. The protein has endonuclease activity, and cleaves the strands at exactly the same time. The cleavage is symmetrical, and gives two recombined DNA molecules with single stranded breaks.[4]
Eukaryotes
The eukaryotic mechanism is much more complex involving different and additional proteins, but follows the same general path.[2] Rad54, a highly conserved eukaryotic protein, is reported to oligomerize on Holliday junctions to promote branch migration.[5]
Archaea
A helicase (designated Saci-0814) isolated from the thermophilic crenarchaeon Sulfolobus acidocaldarius dissociated DNA Holliday junction structures, and showed branch migration activity in vitro.[6] In a S. acidocaldarius strain deleted for Saci-0814, the homologous recombination frequency was reduced five-fold compared to the parental strain indicating that Saci-0814 is involved in homologous recombination in vivo. Based on this evidence it appears that Saci-0814 is employed in homologous recombination in S. acidocaldarius and functions as a branch migration helicase.[6] Homologous recombination appears to be an important adaptation in hyperthermophiles, such as S. acidocaldarius, for efficiently repairing DNA damage. Helicase Saci-0814 is classified as an aLhr1 (archaeal long helicase related 1) under superfamily 2 helicases, and its homologs are conserved among the archaea.
Control
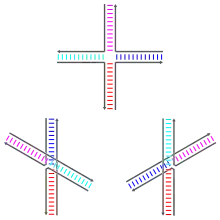
The rate of branch migration is dependent on the amount of divalent ions, specifically magnesium ions (Mg2+), present during recombination.[1] The ions determine which structure the Holliday junction will adopt, as they play a stabilizing role. When the ions are absent, the backbones repel each other and the junction takes on the open X structure.[7] In this condition, migration is optimal and the junction will be free to move up and down the strands.[3] When the ions are present, they neutralize the negatively charged backbone. This allows the strands to move closer together and the junction adopts the stacked X structure.[7] It is during this state that resolution will be optimal, allowing RuvC to bind to the junction.[3]
References
- Lilley, David M. J. (2000-05-01). "Structures of helical junctions in nucleic acids". Quarterly Reviews of Biophysics. 33 (2): 109–159. doi:10.1017/s0033583500003590. ISSN 1469-8994. PMID 11131562. S2CID 40501795.
- "Genetic Recombination | Learn Science at Scitable". www.nature.com. Retrieved 2015-11-13.
- Yamada, Kazuhiro; Ariyoshi, Mariko; Morikawa, Kosuke (2004-04-01). "Three-dimensional structural views of branch migration and resolution in DNA homologous recombination". Current Opinion in Structural Biology. 14 (2): 130–137. doi:10.1016/j.sbi.2004.03.005. PMID 15093826.
- Górecka, K. M.; Komorowska, W.; Nowotny, M. (2013). "Crystal structure of RuvC resolvase in complex with Holliday junction substrate". Nucleic Acids Research. 41 (21): 9945–9955. doi:10.1093/nar/gkt769. PMC 3834835. PMID 23980027.
- Goyal N, Rossi MJ, Mazina OM, Chi Y, Moritz RL, Clurman BE, Mazin AV (2018). "RAD54 N-terminal domain is a DNA sensor that couples ATP hydrolysis with branch migration of Holliday junctions". Nat Commun. 9 (1): 34. Bibcode:2018NatCo...9...34G. doi:10.1038/s41467-017-02497-x. PMC 5750232. PMID 29295984.
- Suzuki, Shoji; Kurosawa, Norio; Yamagami, Takeshi; Matsumoto, Shunsuke; Numata, Tomoyuki; Ishino, Sonoko; Ishino, Yoshizumi (2021). "Genetic and Biochemical Characterizations of aLhr1 Helicase in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius". Catalysts. 12: 34. doi:10.3390/catal12010034.
- Clegg, R. M. (1993-01-01). "The Structure of the Four-Way Junction in DNA". Annual Review of Biophysics and Biomolecular Structure. 22 (1): 299–328. doi:10.1146/annurev.bb.22.060193.001503. PMID 8347993.