C20orf27
UPF0687 protein C20orf27 is a protein that in humans is encoded by the C20orf27 gene.[5][6] It is expressed in the majority of the human tissues. One study on this protein revealed its role in regulating cell cycle, apoptosis, and tumorigenesis via promoting the activation of NFĸB pathway.[7]
| C20orf27 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | C20orf27, chromosome 20 open reading frame 27 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1914576 HomoloGene: 41660 GeneCards: C20orf27 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
The UPF0687 Protein C20orf27 has four other aliases, Chromosome 20 Open Reading Frame 27,[8] Hypothetical Protein LOC54976,[9] C20orf27, and FLJ20550. It is located on the minus strand at 20p13.[8] It consists of 7 exons and 12 introns. This most updated annotation shows that gene C20orf27 starts at 3,753,499 bp to 3,768,388 bp on Chromosome 20.
Transcription
Known isoforms
The C20orf27 gene has 5 transcript isoforms, C20orf27 transcript variant 1, C20orf27 transcript variant 2, C20orf27 transcript variant 3, and C20orf27 transcript variant 4.[8]
Transcript variant 1 encodes for the longest protein isoform, with a size of 1327 bases and 6 exons.[10]
Transcript variant 2 maintains the reading frames and 6 exons compared to transcript variant 1, but it has an alternative spliced site in the coding region.[8] It has a size of 1252 bases.[11]
Transcription variant 3 has a size of 1706 bases and 6 exons.[12] This variant has an alternative spliced site in the coding region and differs in the 5’ UTR, but it still maintains the reading frame seen in transcript variant 1.[8] Despite their differences in size, variant 2 and variant 3 encodes the same protein isoform and this second protein isoform is shorted than the protein isoform encoded by transcript variant 1.
Transcript variant 4 has a size of 1457 bases with 6 exons.[13] Compared to variant 1, it uses an alternative 5’-most exon and an alternative splice site.[8] Because of the presence of an upstream ORF that is predicted to interfere with translation of this variant, the transcription variant 4 does not encode any protein.
The information on transcript variant X1 comes from GRCh38.p13 Primary Assembly.[14] This variant has a size of 1195 bases, and the number of exons in this variant remains unknown.
Proteins
Physical features

The human gene C20orf27 has three known isoforms.[8]
Isoform 1 has 199 amino acid residues and a domain named DUF4517. Isoform 2 has 174 amino acid residues, and isoform X1 has 154 amino acid residues. All three isoforms contain the same domain DUF4517. The function of domain DUF4517 requires future research.
The predicted isoelectric point of unmodified protein C20orf27 is 6.89.[15]
The percentage of each amino acid residue is about its average percentage among human proteins.[16] Overall, the positively charged amino acid residues in human protein C20orf27 outnumbers the negatively charged amino acid residues. Protein C20orf27 has no high scoring hydrophobic regions, no highly charged regions, and no transmembrane regions.
SPAS predicts two repetitive structures. The first repetitive structure is amino acid alphabet structures with a core block length of 4. The total number of this structure in human protein C20orf27 is 15. The second repetitive structure is an 11-letter reduced alphabet structure with a core block length of 8. This charged alphabet structure predicts to appear 8 times in human protein C20orf27. There are no predicted clusters of amino acid multiples.
Post-translation modifications
The predicted molecular weight of C20orf27 is 21.6 kDa.[16] A Western Blot binding pattern on protein C20orf27 with its polyclonal antibody reveals that the experimental molecular weight of protein C20orf27 is about 22 kDa.[17] This suggests that there are relatively few post-translation modifications on protein C20orf27.
There is no predicted signal peptide or cleavage site.

There are many predicted phosphorylation sites along the sequence of protein C20orf27, including four sites for protein kinase A (PKA), two sites for protein kinase C (PKC), three sites for casein kinase 2 (CKII), one site for ribosomal S6 kinase (RSK), one site for cGMP-dependent protein kinase or Protein Kinase G (PKG), and one site for ataxia-telangiectasia mutated (ATM) serine/threonine protein kinase.[18]
Protein C20orf27 is predicted to have other post-translation modification sites including five palmitoylation sites,[19] one c-mannosylation site,[20] and two sumoylation sites.[21]
Structure
Three stretches of beta sheet from amino acid 62 to 67, 76 to 87, and 92 to 100 are predicted with the highest confidence using CFSSP[22] and Phyre2.[23] A model predicted by I-TASSER[24] shows that the tertiary structure of human protein C20orf27 is a combination of many beta sheets. This confirms the predictions made by CFSSP and Phyre2.
Expression
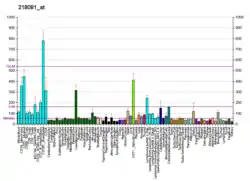
Protein C20orf27 is expressed ubiquitously throughout different human tissues. Microarray-assessed tissue expression pattern suggests caudate nucleus has the highest expression of protein C20orf27.[27]
Other than caudate nucleus, protein C20orf27 expression measure ranks at the top 25% among 100 proteins in pons, fetal brain, BM- CD105+ endothelial, BM- CD34+, bone marrow, adipocyte, uterus corpus, 721 BLymphoblast, PB- CD56+NK cells, BM- CD33+ myeloid, colorectal adenocarcinoma, leukemia chronic Myelogenous K-562, leukemia lymphoblastic (MOLT-4), and leukemia promyelocytic-HL-60.
In situ hybridization data has shown that the expression of C20orf27 in airway epithelial cells (AECs) can be correlated to chronic lung diseases.[28] After AECs are treated with IL-13, which is a cytokine expressed by CD4 T helper cells, AECs begin to secrete excess mucous, and excess mucous secretion in the airway is a mark of chronic lung diseases.
Regulation of expression
Gene level expression
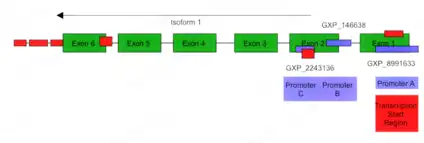
There are three promoter regions in gene C20orf27.
Five transcription factors that bind to the promoter region of gene C20orf27[29] have been discovered, including MITF, JUN, ZNF282, FOXA1, and TCF7L2.
Using genomatix, more transcription factor binding sites are predicted.[30] Transcription binding matrix, like EGR/nerve growth factor induced protein C & related factors, GC-Box factors SP1/GC, Krueppel like transcription factors, Myc associated zinc fingers, vertebrate homologues of enhancer of split complex, E-box binding factors, E2F-myc activator/cell cycle regulator, and BED subclass of zinc-finger proteins, are predicted to give the highest matrix similarity.
Transcript level regulation

Predicted miRNA binding sites in 3' end of C20orf27 mRNA which sequences are also conserved evolutionarily are hsa-miR-7856-5p, hsa-miR-671-5p, hsa-miR-4768, hsa-miR-6791-3p, hsa-miR-6829-3p, hsa-miR-548d-3p, hsa-miR-548-3p, hsa-miR-548z, and hsa-miR-548h-3p.[31]
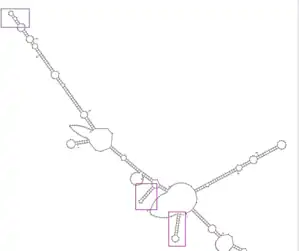
The formation of three stem loops is conserved in different predicted models.[32] The three stem loops start from the 5' end of C20orf27 mRNA base 1 to base 27, base 56 to base 74, and base 116 to base 130.
The mRNA of C20orf27 has about 23 predicted mRNA binding protein binding sites which sequences are also conserved in evolution.[33] The names of these mRNA binding proteins are BRUNOL5, BRUNOL6, PCBP2, TARDBP, MBNL1, CUG-BP, PCBP3, PTBP1, RBM5, SRSF1, HNRNPH2, FMR1, HNRNPF, LIN28A, CPEB4, HNRNPC, HNRNPCL1, HNRNPM, HuR, RALY, PABPC1, PABPC4, SART3, and SRSF10.
Function and clinical significance
Interacting proteins
Interactors of protein C20orf27 found in Y2H screen are replicase polyprotein 1ab from coronavirus,[34] RAIYL,[35] PHKB,[35] FERMT2[36] from human. The function of replicase polyprotein 1ab is transcribing and replicating viral RNAs, and it contains the proteinases responsible for the cleavages of the polyprotein.[37] The function of RAIYL,[35] PHKB,[35] and FERMT2[36] remain unknown.
Other interactors that discovered by pull-down assays include PPP1CA,[38] PPP1CC,[38] PPP1CB,[39] PPP1R7,[39] PSME3,[40] RBFOX2,[40] and DMWD.[40] Interactors PPP1CA, PPP1CB, PPP1CC, and PPP1R7 have similar functions. They involve in the regulation of a variety of cellular processes, such as cell division, glycogen metabolism, muscle contractility, protein synthesis, and HIV-1 viral transcription.[41][42][43][44] PSME3 facilitates the MDM2-p53/TP53 interaction which promotes ubiquitination- and MDM2-dependent proteasomal degradation of p53/TP53, limiting its accumulation and resulting in inhibited apoptosis after DNA damage, and might play a role in cell cycle regulation.[45][46][47][48][49][50][51] RBFOX2 regulates alternative splicing events by binding to 5'-UGCAUGU-3' elements.[52] The function of DMWD is unknown.
The above evidence suggests protein C20orf27 plays a role in cell cycle regulation, cell proliferation and differentiation, and cell survival.
Clinical significance
Human protein C20orf27 and its variants have not been discovered to be associated with any diseases or disorders.
Homology and evolution history
Paralogs
There are no known paralogs.[8]
Orthologs
There are about 281+ known orthologs for this gene, ranging from primates to invertebrates.[8]

The closest related orthologs are selected from primates and mammals, and the sequence similarity ranks from 75% to 100%. The moderately related orthologs are selected from fishes and birds, and the sequence similarity ranks from 55% to 75%. The most distantly related orthologs are selected from invertebrates and trichoplax, and the sequence similarity ranks from 40% to 55%. The conserved amino acids are bold in the conceptional translation diagram.
| Gene name | Genus and Species | taxonomic group | Common Names | Accession Num. | Protein Length | Seq Identity | Seq Similarity | MYA |
| C20orf27 | Homo sapiens | Primates | Human | NP_001034229.1 | 199 aa | 100% | 100% | 0 |
| C20orf27 | Macaca mulatta | Primates | Old World Monkey | AFE71948.1 | 197 aa | 98.50% | 99% | 29.44 |
| C20orf27 | Mus musculus | Rodentia | House mouse | NP_001298067.1 | 177 aa | 79.40% | 82.90% | 90 |
| C20orf27 | Rhinolophus ferrumequinum | Chiroptera | Greater horseshoe bat | XP_032951391.1 | 174 aa | 82.40% | 85.40% | 96 |
| C20orf27 | Condylura cristata | Eulipotyphla | Star-nosed mole | XP_012583921.1 | 184 aa | 76.60% | 79.90% | 96 |
| C20orf27 | Dromaius novaehollandiae | Casuariiformes | Emu | XP_025975497.1 | 174 aa | 65.30% | 72.90% | 312 |
| C20orf27 | Gopherus evgoodei | Testudines | Gopher Tortoises | XP_030419106.1 | 176 aa | 61.80% | 73.90% | 312 |
| C20orf27 | Strigops habroptila | Psittaciformes | Kakapo | XP_030348224.1 | 174 aa | 59.30% | 70.40% | 312 |
| C20orf27 | Thamnophis elegans | Scaled reptiles | Western terrestrial garter snake | XP_032094251.1 | 174 aa | 56.80% | 68.30% | 312 |
| C20orf27 | Taeniopygia guttata | Passeriformes | Zebra finch | NP_001232719.1 | 176 aa | 56.70% | 67.50% | 312 |
| C20orf27 | Xenopus tropicalis | Frogs | Western clawed frog | NP_001007504.1 | 174 aa | 59.30% | 72.40% | 351.8 |
| C20orf27 | Scophthalmus maximus | Pleuronectiformes | Turbot | AWP06390.1 | 179 aa | 29.70% | 43.20% | 435 |
| C20orf27 | Callorhinchus milii | Chimaera | Australian ghostshark | XP_007906148.1 | 179 aa | 54.00% | 63.90% | 473 |
| C20orf27 | Petromyzon marinus | Petromyzontiformes | Sea lamprey | XP_032806447.1 | 173 aa | 47.60% | 55.80% | 615 |
| C20orf27 | Anneissia japonica | Comatulida | comasterids | XP_033124803.1 | 184 aa | 26.60% | 46.30% | 684 |
| C20orf27 | Ixodes scapularis | Ixodida | Deer tick | XP_002403181.1 | 165 aa | 28.80% | 44.20% | 797 |
| C20orf27 | Limulus polyphemus | Xiphosura | Atlantic horseshoe crab | XP_022257482.1 | 173 aa | 28.80% | 44.50% | 797 |
| C20orf27 | Crassostrea gigas | Ostreida | Pacific oyster | XP_011438297.1 | 162 aa | 24.90% | 44.00% | 797 |
| C20orf27 | Drosophila subobscura | Fly | Fruit Fly | XP_034657203.1 | 179 aa | 24.70% | 37.70% | 797 |
| C20orf27 | Nematostella vectensis | Sea anemone | Starlet sea anemone | XP_001627979.1 | 169 aa | 30.30% | 43.30% | 824 |
| C20orf27 | Trichoplax | Trichoplax | Trichoplax | RDD38604.1 | 166 aa | 22.3% | 40.0% | 1017 |
References
- GRCh38: Ensembl release 89: ENSG00000101220 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000027327 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Deloukas P, Matthews LH, Ashurst J, Burton J, Gilbert JG, Jones M, et al. (Jan 2002). "The DNA sequence and comparative analysis of human chromosome 20". Nature. 414 (6866): 865–71. Bibcode:2001Natur.414..865D. doi:10.1038/414865a. PMID 11780052.
- "Entrez Gene: C20orf27 chromosome 20 open reading frame 27".
- Gao J, Wang Y, Zhang W, Zhang J, Lu S, Meng K, et al. (February 2020). "C20orf27 Promotes Cell Growth and Proliferation of Colorectal Cancer via the TGFβR-TAK1-NFĸB Pathway". Cancers. 12 (2): 336. doi:10.3390/cancers12020336. PMC 7072304. PMID 32024300.
- "C20orf27 chromosome 20 open reading frame 27 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-07-31.
- "C20orf27 Gene - GeneCards | CT027 Protein | CT027 Antibody". www.genecards.org. Retrieved 2020-08-05.
- "Homo sapiens chromosome 20 open reading frame 27 (C20orf27), transcript variant 1, mRNA". 2020-05-12.
{{cite journal}}: Cite journal requires|journal=(help) - "Homo sapiens chromosome 20 open reading frame 27 (C20orf27), transcript variant 2, mRNA". 2020-07-10.
{{cite journal}}: Cite journal requires|journal=(help) - "Homo sapiens chromosome 20 open reading frame 27 (C20orf27), transcript variant 3, mRNA". 2020-05-12.
{{cite journal}}: Cite journal requires|journal=(help) - "Homo sapiens chromosome 20 open reading frame 27 (C20orf27), transcript variant 4, non-coding RNA". 2020-02-14.
{{cite journal}}: Cite journal requires|journal=(help) - "PREDICTED: Homo sapiens chromosome 20 open reading frame 27 (C20orf27), transcript variant X1, mRNA". 2020-05-28.
{{cite journal}}: Cite journal requires|journal=(help) - ExPASy Bioinformatics Resource Portal entry on Compute pl/Mw tool https://web.expasy.org/compute_pi/. Retrieved 2020-7-31.
- EMBL-EBI (European Bioinformatics Institute) entry on Statistical Analysis of Protein Sequences (SAPS) tool https://www.ebi.ac.uk/Tools/seqstats/saps/. Retrieved 2020-7-31.
- "C20orf27 Antibody (PA5-61529)". www.thermofisher.com. Retrieved 2020-07-31.
- ExPASy Bioinformatics Resource Portal entry on NetPhos 3.1 Serve. http://www.cbs.dtu.dk/services/NetPhos/. Retrieved 2020-7-31.
- GSS-Palm. Prediction of Palmitoylation Site. http://csspalm.biocuckoo.org/. Retrieved on 2020-7-31.
- NetCGlyc 1.0. Neural network predictions of C-mannosylation sites in mammalian proteins. http://www.cbs.dtu.dk/services/NetCGlyc/. Retrieved on 2020-7-31.
- GPS-SUMO. Prediction of SUMOylation Sites & SUMO-binding Motifs. http://sumosp.biocuckoo.org/ Retrieved on 2020-7-31.
- CFSSP: Chou and Fasman Secondary Structure Prediction server. http://www.biogem.org/tool/chou-fasman/. Retrieved 2020-8-01.
- Phyre2: Protein Homology/analogY Recognition Engine V 2.0. http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index. Retrieved on 2020-8-01.
- I-TASSER (Iterative Threading ASSEmbly Refinement. A server for protein structure & function prediction. https://zhanglab.ccmb.med.umich.edu/I-TASSER/. Retrieved on 2020-8-01.
- ThermoFisher entry on C20orf27 Polyclonal Antobody. Retrieved on 2020-08-02.
- PSORT: a portal to protein subcellular localization resources. Retrieved on 2020-8-02.
- NCBI (National Center for Biotechnology Information) GEO Profile entry on C20orf27 gene https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS596:50314_i_at. Retrieved on 2020-8-01.
- NCBI (National Center for Biotechnology Information) GEO Profile entry on C20orf27 gene. Retrieved on 2020-8-01.
- SPP (The Signaling Pathway Project) transcription factors report on C20orf27 gene. Retrieved on 2020-8-01.
- "Genomatix - NGS Data Analysis & Personalized Medicine". www.genomatix.de. Retrieved 2020-08-02.
- miRDB. Online database for miRNA targeting prediction and functional annotations. Retrieved on 2020-8-01.
- The mfold web server. Web server for nucleic acid folding and hybridization prediction. Retrieved on 2020-8-01.
- RBPmap: mapping binding sites of RNA binding proteins. Retrieved on 2020-8-02.
- Pfefferle S, Schöpf J, Kögl M, Friedel CC, Müller MA, Carbajo-Lozoya J, et al. (October 2011). "The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors". PLOS Pathogens. 7 (10): e1002331. doi:10.1371/journal.ppat.1002331. PMC 3203193. PMID 22046132.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Sügis E, Dauvillier J, Leontjeva A, Adler P, Hindie V, Moncion T, et al. (August 2019). "HENA, heterogeneous network-based data set for Alzheimer's disease". Scientific Data. 6 (1): 151. Bibcode:2019NatSD...6..151S. doi:10.1038/s41597-019-0152-0. PMC 6694132. PMID 31413325.
- Lokugamage KG, Narayanan K, Huang C, Makino S (December 2012). "Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation". Journal of Virology. 86 (24): 13598–608. doi:10.1128/JVI.01958-12. PMC 3503042. PMID 23035226.
- Yadav L, Tamene F, Göös H, van Drogen A, Katainen R, Aebersold R, et al. (April 2017). "Systematic Analysis of Human Protein Phosphatase Interactions and Dynamics". Cell Systems. 4 (4): 430–444.e5. doi:10.1016/j.cels.2017.02.011. PMID 28330616.
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, et al. (May 2016). "An organelle-specific protein landscape identifies novel diseases and molecular mechanisms". Nature Communications. 7 (1): 11491. Bibcode:2016NatCo...711491B. doi:10.1038/ncomms11491. PMC 4869170. PMID 27173435.
- "C20orf27 protein (human) - STRING interaction network". string-db.org. Retrieved 2020-08-02.
- Mi J, Guo C, Brautigan DL, Larner JM (February 2007). "Protein phosphatase-1alpha regulates centrosome splitting through Nek2". Cancer Research. 67 (3): 1082–9. doi:10.1158/0008-5472.CAN-06-3071. PMID 17283141.
- Nie H, Zheng Y, Li R, Guo TB, He D, Fang L, et al. (March 2013). "Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis". Nature Medicine. 19 (3): 322–8. doi:10.1038/nm.3085. PMID 23396208. S2CID 7100407.
- Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q, et al. (2015). "ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation". Autophagy. 11 (8): 1308–25. doi:10.1080/15548627.2015.1060386. PMC 4590681. PMID 26083323.
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, et al. (January 2015). "Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres". Developmental Cell. 32 (1): 68–81. doi:10.1016/j.devcel.2014.11.030. PMID 25556658.
- Realini C, Jensen CC, Zhang Z, Johnston SC, Knowlton JR, Hill CP, Rechsteiner M (October 1997). "Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators". The Journal of Biological Chemistry. 272 (41): 25483–92. doi:10.1074/jbc.272.41.25483. PMID 9325261. S2CID 83200535.
- Wilk S, Chen WE, Magnusson RP (November 2000). "Properties of the nuclear proteasome activator PA28gamma (REGgamma)". Archives of Biochemistry and Biophysics. 383 (2): 265–71. doi:10.1006/abbi.2000.2086. PMID 11185562.
- Li J, Gao X, Joss L, Rechsteiner M (June 2000). "The proteasome activator 11 S REG or PA28: chimeras implicate carboxyl-terminal sequences in oligomerization and proteasome binding but not in the activation of specific proteasome catalytic subunits". Journal of Molecular Biology. 299 (3): 641–54. doi:10.1006/jmbi.2000.3800. PMID 10835274.
- Magni M, Ruscica V, Buscemi G, Kim JE, Nachimuthu BT, Fontanella E, et al. (December 2014). "Chk2 and REGγ-dependent DBC1 regulation in DNA damage induced apoptosis". Nucleic Acids Research. 42 (21): 13150–60. doi:10.1093/nar/gku1065. PMC 4245943. PMID 25361978.
- Li J, Gao X, Ortega J, Nazif T, Joss L, Bogyo M, et al. (July 2001). "Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta". The EMBO Journal. 20 (13): 3359–69. doi:10.1093/emboj/20.13.3359. PMC 125523. PMID 11432824.
- Gao X, Li J, Pratt G, Wilk S, Rechsteiner M (May 2004). "Purification procedures determine the proteasome activation properties of REG gamma (PA28 gamma)". Archives of Biochemistry and Biophysics. 425 (2): 158–64. doi:10.1016/j.abb.2004.03.021. PMID 15111123.
- Zhang Z, Zhang R (March 2008). "Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation". The EMBO Journal. 27 (6): 852–64. doi:10.1038/emboj.2008.25. PMC 2265109. PMID 18309296.
- Norris JD, Fan D, Sherk A, McDonnell DP (March 2002). "A negative coregulator for the human ER". Molecular Endocrinology. 16 (3): 459–68. doi:10.1210/mend.16.3.0787. PMID 11875103.
External links
- Human C20orf27 genome location and C20orf27 gene details page in the UCSC Genome Browser.
Further reading
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J (May 2003). "Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides". Nature Biotechnology. 21 (5): 566–9. doi:10.1038/nbt810. PMID 12665801. S2CID 23783563.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.