C5orf49
Chromosome 5 open reading frame forty-nine, also known as C5orf49, is a protein that in humans is encoded by the C5orf49 gene. Aliases for C5orf49 include Chromosome 5 Open Reading Frame 49, Uncharacterized Protein C5orf49 and LOC134121.[5] C5orf49 is predicted to localize to the cilia and have ciliary functions.[6]
| C5orf49 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | C5orf49, chromosome 5 open reading frame 49 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1916565 HomoloGene: 28246 GeneCards: C5orf49 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene

C5orf49 is found on chromosome 5, cytoband p15 between base pairs 7,830,378 and 7,851,151, meaning it has a length of 20,774 base pairs.[7] This gene has two splice forms, one that is 147 amino acids in length and another that is 145 amino acids in length.[8] C5orf49 is oriented on the minus strand.[5] Neighboring genes of C5orf49 include, FASTKD3, MTRR, and ADCY2.
Promoter
C5orf49 has one upstream promoter, GXP_1271072, that regulates both of the primary transcripts.[8] GXP_1271072 is 1,396 base pairs in length, spanning from base pair 7,851,094 to base pair 7,852,489 on chromosome 5. The transcription start region for the longest transcript of 147 amino acids spans from base pair 7,851,148 to base pair 7,851,164 on chromosome 5.
Protein
Structure

C5orf49 is characterized by the presence of the protein domain DUF4541.[5] Within this protein domain, there is a conserved KLHRDDR sequence motif and a single completely conserved residue Y that may be functionally important.[9] Domain is shown on the annotated conceptual translation.
Predicted properties
The following properties of C5orf49 were predicted using bioinformatic analysis:
- Molecular Weight: 17 kDa[5]
- Isoelectric point: 7.0[10]
- Post-translational modification: fourteen post-translational modifications are predicted:
- Seven phosphorylation sites at positions 8, 9, 11, 80, 100, 135, and 147 on the protein sequence[11]
- Six ubiquitination sites at 16, 39, 69, 104, 137.
- Two acetylation sites at 39 and 104.
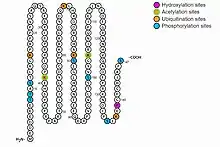 Post-translational modifications for C5orf49
Post-translational modifications for C5orf49
Tissue distribution

Expression data indicate expression most significantly in the lung, brain, and spinal cord tissues.[12]
Binding partners
CDKN2d, HSF2BP, KRT31 and KRT34 were found to be binding partners of C5orf49 by two hybrid prey pooling approach and two hybrid array.[13]
Species Distribution

C5orf49 shows conservation through mammals and orthologs can be found in flatworms and sea anemone. The table to the right shows a spread of some orthologs found using BLAST.[14] C5orf49 is not found in sponges, which diverged at a median date of 777 million years ago (MYA),[15] and it is found in its most distant ortholog 736 MYA. Therefore, C5orf49 diverged as a gene between 777 MYA and 736 MYA.
Evolution
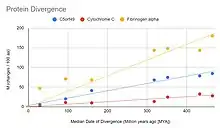
C5orf49 does not show a fast or slow evolution rate over time when compared to cytochrome C and fibrinogen alpha. This is shown by the protein divergence graph on the right.
References
- GRCh38: Ensembl release 89: ENSG00000215217 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000021534 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "C5orf49". GeneCards: Human Gene Database. Archived from the original on 2011-09-01.
- Sigg, Monika Abedin; Menchen, Tabea; Lee, Chanjae; Johnson, Jeffery; Jungnickel, Melissa K.; Choksi, Semil P.; Garcia, Galo; Busengdal, Henriette; Dougherty, Gerard; Pennekamp, Petra; Werner, Claudius (2017-12-18). "Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways". Developmental Cell. 43 (6): 744–762.e11. doi:10.1016/j.devcel.2017.11.014. ISSN 1534-5807. PMC 5752135. PMID 29257953.
- "C5orf49 chromosome 5 open reading frame 49 [Homo sapiens (human)] – Gene – NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-12-18.
- "Genomatix: ElDorado entry on C5orf49". Genomatix Software Suite.
- "InterPro". www.ebi.ac.uk. Retrieved 2021-12-18.
- "C5orf49 (human)". www.phosphosite.org. Retrieved 2021-12-18.
- Wang, D (2020). "MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization". Nucleic Acids Research. 48 (W1): W140–W146. doi:10.1093/nar/gkaa275. PMC 7319475. PMID 32324217.
- "Home – GEO – NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-12-18.
- "IntAct Portal". www.ebi.ac.uk. Retrieved 2021-12-18.
- "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2021-12-18.
- "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 2021-12-18.