CCDC188
CCDC188 or coiled-coil domain containing protein is a protein that in humans is encoded by the CCDC188 gene.[4]
| CCDC188 | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||
| Aliases | CCDC188, coiled-coil domain containing 188 | ||||||||||||||||||||||||||||||
| External IDs | HomoloGene: 90442 GeneCards: CCDC188 | ||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
Gene
Human CCDC188 gene spans 3715 nucleotides and is located on the minus strand of chromosome 22 at 22q11.21.[5] It is a protein coding gene that encodes CCDC188 protein.[6] The mRNA transcript consists of 9 different exons which are spliced to form the 6 distinct CCDC188 protein isoforms.[7] Genetic neighbors of CCDC188 include ZDHHC8, SNORA77B, and RANBP1.
| Transcript Name | Accession Number | Exons | Nucleotide length | Final Protein Length (aa) |
|---|---|---|---|---|
| CCDC188 | NM_001365892.2 | 9 | 1476 | 402 |
| CCDC188 Isoform X1 | XM_005261238.3 | 7 | 2501 | 435 |
| CCDC188 Isoform X2 | XM_005261239.3 | 7 | 2445 | 416 |
| CCDC188 Isoform X4 | XM_011530170.2 | 9 | 2364 | 402 |
| CCDC188 Isoform X5 | XM_011530171.2 | 7 | 2396 | 400 |
| CCDC188 Isoform X6 | XM_005261241.3 | 7 | 2393 | 399 |
RNA expression
CCDC188 is expressed at low levels across all adult tissues with increased expression in the pituitary gland and testis.[8] CCDC188 has decreased expression in G1 of the cell cycle.

Genes with similar mRNA expression in the hypothalamus, supraoptic nucleus, and dentate gyrus are shown in the table below.

| Structure | Gene | Expande Name | Function | Pearson Coefficient |
|---|---|---|---|---|
| Hypothalamus | SKAP1 | Src Kinase Associated Phosphoprotein 1 | Couple T-cell antigen receptor stimulation to the activation of integrins | 0.71 |
| CLIC1 | Chloride Intracellular Channel 1 | Nuclear chloride ion channel activity | 0.659 | |
| PPAPDC1B | Phospholipid Phosphatase 5 | Converts diacylglycerol pyrophosphate into phosphatidate | 0.656 | |
| FAM27A | NA | lncRNA | 0.653 | |
| ZCWPW2 | Zinc Finger CW-Type and PWWP Domain Containing 2 | Transcription factor that binds to histone methyl groups | -0.612 | |
| ZNF519 | Zinc Finger Protein 519 | Transcription Factor | -0.61 | |
| SLC8A1 | Solute Carrier Family 8 Member A1 | Calcium and sodium ion exchange mediator | -0.601 | |
| Supraoptic Nucleus | ZNF181 | Zinc Finger Protein 181 | Transcription Factor | 0.996 |
| DYNC1LI1 | Dynein Cytoplasmic 1 Light Intermediate Chain | Intracellular trafficking and chromosome segregation during mitosis | 0.991 | |
| COX18 | Cytochrome C Oxidase Assembly Factor 18 | Integral membrane insertion into inner mitochondrial membrane | 0.991 | |
| LAMA2 | Laminin Subunit Alpha 2 | Attachment to basement membrane | 0.991 | |
| KCTD8 | Potassium Channel Tetramerization Domain Containing 8 | Determines kinetics of GABA-B receptor | -0.985 | |
| KLHL2 | Kelch Like Family Member 2 | Mediates ubiquitination of target proteins | -0.985 | |
| Dentate Gyrus | CXCL9 | CXC Motif Chemokine Ligand 9 | Antimicrobial protein | 0.879 |
| KYNU | Kynureninase | Biosynthesis of NAD cofactors from tryptophan | 0.866 | |
| MASP1 | Mannose-Binding Lectin Associated Serine Protease 1 | Serine protease essential for adaptive immune response | 0.808 | |
| TRPC6 | Transient Receptor Potential Cation Channel | Receptor activated calcium channel | 0.805 |
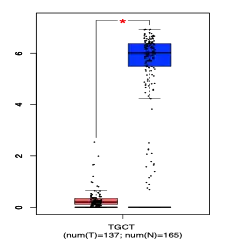
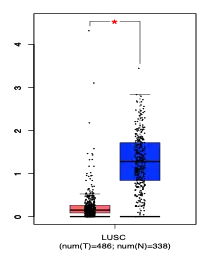
The promoter region for CCDC188 contains highly conserved p53[9] and CREB-ATF4[10] binding sites.[11] Chromatin-immunoprecipitation analysis confirms p53 binding to the promoter region of CCDC188.[12] Significantly repressed CCDC188 mRNA expression is found in both testicular germ line tumors and lung squamous cell cancer.[13]
Copy number variations of CCDC188 have also been identified in lung squamous cell tumors with 16 tumors having amplifications and 4 having homodeletions.[14] Genes with significantly increased mRNA expression under CCDC188 amplification in lung squamous cell tumors are shown in the table below.
| Gene | p-Value | q-Value | Genetic Locus |
|---|---|---|---|
| MAGED1 | 3.23E-06 | 0.013 | Xp11.22 |
| MAPK13 | 5.37E-06 | 0.0149 | 6p21.31 |
| RBM4 | 9.36E-06 | 0.0202 | 11q13.2 |
| NYNRIN | 3.04E-05 | 0.0328 | 14q12 |
| HDAC7 | 7.1E-05 | 0.0486 | 12q13.11 |
| ZNF675 | 7.25E-05 | 0.0486 | 19p12 |
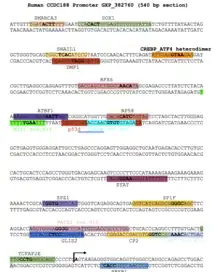
Other predicted transcription factor binding sites for CCDC188 are shown in the figure to the right.[15]
Transcript regulation
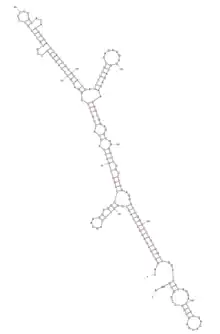
Predicted CCDC188 3'UTR stem loops are shown in the figure below.[16]
Protein
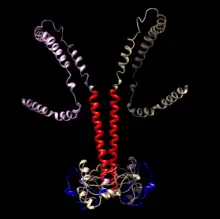
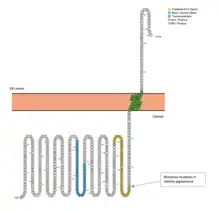
CCDC188 protein is 402 amino acids long and is 4.3 kDa.[17] The protein contains a leucine zipper and transmembrane domain.[18] The presence of both a leucine zipper domain and transmembrane domain suggests that CCDC188 protein functions as a transcription factor that is tightly regulated and must be cleaved out of a membrane to be activated. The inactive form of the protein is predicted to be located in the endoplasmic reticulum with the N-terminus and basic leucine zipper oriented in the cytosol.[19] Other membrane bound basic leucine zippers include ATF6 and OASIS.[20] Known nuclear transportation routes for membrane bound transcription factors in the endoplasmic reticulum include ubiquitination and destruction of the ER lumen region and COPII vesicular transport to the Golgi for proteolytic cleavage by resident proteases.[21]
Post-translational modifications
Two phosphate groups have been experimentally verified on serine residues 322 and 324 in B-cell leukemia.[22]
Homology
CCDC188 is conserved throughout all mammals including monotremes, marsupials, and placentals[23]
| Clade | Genus & Species | Common Name | Taxonomic Group | Divergence Date (MYA) | Accession Number | Query Cover | Sequence Length (aa) | Sequence Identity (%) | Sequence Similarity (%) |
|---|---|---|---|---|---|---|---|---|---|
| Placentals | Homo sapiens | Human | Primate | 0 | NP_001352821.1 | 100 | 402 | 100 | 100 |
| Gorilla gorilla | Western Gorilla | Primate | 9 | XP_004063092.3 | 100 | 402 | 97 | 98 | |
| Rhinopithecus roxellana | Golden Snub Nosed Monkey | Primate | 29 | XP_010386733.2 | 100 | 393 | 90 | 91 | |
| Marmota flaviventris | Yellow-Bellied Marmot | Rodentia | 89 | XP_027780043.1 | 100 | 407 | 76 | 82 | |
| Leptonychotes weddelli | Weddell Seal | Carnivora | 94 | XP_030873069.1 | 100 | 407 | 76 | 82 | |
| Ailuropoda melanoleuca | Giant Panda | Carnivora | 94 | XP_011225007.2 | 100 | 407 | 76 | 82 | |
| Canis lupus | Grey Wolf | Carnivora | 94 | XP_025330588.1 | 100 | 407 | 76 | 82 | |
| Talpa occidentalis | Spanish Mole | Insectivora | 94 | XP_037351914.1 | 100 | 406 | 74 | 79 | |
| Globicephala melas | Long Finned Pilot Whale | Delphinidae | 94 | XP_030692560.1 | 100 | 408 | 74 | 80 | |
| Molossus molossus | Velvety Free-Tailed Bat | Chiroptera | 94 | XP_036132060.1 | 100 | 404 | 74 | 79 | |
| Eptesicus fuscus | Big Brown Bat | Chiroptera | 94 | XP_008140813.2 | 101 | 404 | 73 | 80 | |
| Rhinolophus ferrumequinum | Greater Horshoe Bat | Chiroptera | 94 | XP_032953151.1 | 100 | 407 | 72 | 79 | |
| Marsupials | Phascolarctos cinereus | Koala | Phascolarctidae | 160 | XP_020852118.1 | 41 | 231 | 44 | 65 |
| Dromiciops gliroides | Colocolo Opossum | Microbiotheridae | 160 | XP_043845525.1 | 62 | 365 | 42 | 61 | |
| Monodelphis domestica | Gray Short Tailed Opossum | Didelphidae | 160 | XP_007490407.1 | 62 | 311 | 41 | 61 | |
| Vombatus ursinus | Common Wombat | Vombatidae | 160 | XP_027703176.1 | 62 | 309 | 40 | 61 | |
| Trichosurus vulpecula | Brushtail Possum | Phalangeroidae | 160 | XP_036604697.1 | 62 | 289 | 40 | 59 | |
| Sarcophilus harrisii | Tasmanian Devil | Dasyuridae | 160 | XP_031804879.1 | 65 | 313 | 38 | 52 | |
| Monotremes | Ornithorhynchus anatinus | Duck-Billed Platypus | Platypus | 180 | XP_028905014.1 | 40 | 246 | 35 | 57 |
| Tachyglossus aculeatus | Short-Beaked Echidna | Echidna | 180 | XP_038618232.1 | 40 | 383 | 35 | 55 |
When CCDC188 first appeared approximately 180 million years ago in monotremes, it lacked a basic leucine zipper. Marsupials were the first mammals to evolve a CCDC188 basic leucine zipper domain. The rate of evolution of CCDC188 measured by sequence identity to humans shows that CCDC188 initially evolved quickly at a rate of 0.97 changes per 100 amino acids per million years. Beginning with the first placentals, CCDC188 evolution slowed to a rate of 0.45 changes per 100 amino acids per million years. One paralog for CCDC188 exists in humans known as CCDC188-like. This gene first appeared in marsupials.

Pathology
A nonsense mutation in the coding region of CCDC188 has been implicated in retinitis pigmentosa,[24] a retinal degeneration process marked by uncontrolled death of rod cells. CCDC188 is also deleted in 22q11.2 deletion syndrome.
References
- GRCh38: Ensembl release 89: ENSG00000234409 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "coiled-coil domain-containing protein 188 [Homo sapiens]". NCBI Protein. Retrieved 2021-09-26.
- "CCDC188 Gene". GeneCards. Retrieved 26 September 2021.
- "CCDC188". NCBI Gene. Retrieved 28 September 2021.
- "CCDC188". NCBI Gene. Retrieved 28 September 2021.
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. (January 2015). "Proteomics. Tissue-based map of the human proteome". Science. 347 (6220): 1260419. doi:10.1126/science.1260419. PMID 25613900. S2CID 802377.
- Fridman JS, Lowe SW (December 2003). "Control of apoptosis by p53". Oncogene. 22 (56): 9030–9040. doi:10.1038/sj.onc.1207116. PMID 14663481. S2CID 16321935.
- Wortel IM, van der Meer LT, Kilberg MS, van Leeuwen FN (November 2017). "Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells". Trends in Endocrinology and Metabolism. 28 (11): 794–806. doi:10.1016/j.tem.2017.07.003. PMC 5951684. PMID 28797581.
- "El Dorado". Genomatix. Intrexon Bioinformatics Germany GmbH 2019. Retrieved 6 December 2021.
- Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, et al. (December 2018). "ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data". EMBO Reports. 19 (12). doi:10.15252/embr.201846255. PMC 6280645. PMID 30413482.
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (July 2017). "GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses". Nucleic Acids Research. 45 (W1): W98–W102. doi:10.1093/nar/gkx247. PMC 5570223. PMID 28407145.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. (May 2012). "The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data". Cancer Discovery. 2 (5): 401–404. doi:10.1158/2159-8290.CD-12-0095. PMC 3956037. PMID 22588877.
- "El Dorado". Genomatix. Intrexon Bioinformatics Germany GmbH 2019. Retrieved 6 December 2021.
- Markham NR, Zuker M (2008). UNAFold: software for nucleic acid folding and hybridization. Methods in Molecular Biology. Vol. 453. pp. 3–31. doi:10.1007/978-1-60327-429-6_1. PMID 18712296.
- "coiled-coil domain-containing protein 188 [Homo sapiens]". NCBI Protein. Retrieved 2021-09-26.
- "coiled-coil domain-containing protein 188 [Homo sapiens]". NCBI Protein. Retrieved 2021-09-26.
- Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O (November 2017). "DeepLoc: prediction of protein subcellular localization using deep learning". Bioinformatics. 33 (21): 3387–3395. doi:10.1093/bioinformatics/btx431. PMID 29036616.
- Stirling J, O'hare P (January 2006). "CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P". Molecular Biology of the Cell. 17 (1): 413–426. doi:10.1091/mbc.e05-06-0500. PMC 1345678. PMID 16236796.
- Liu Y, Li P, Fan L, Wu M (April 2018). "The nuclear transportation routes of membrane-bound transcription factors". Cell Communication and Signaling. 16 (1): 12. doi:10.1186/s12964-018-0224-3. PMC 5883603. PMID 29615051.
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E (January 2015). "PhosphoSitePlus, 2014: mutations, PTMs and recalibrations". Nucleic Acids Research. 43 (Database issue): D512–D520. doi:10.1093/nar/gku1267. PMC 4383998. PMID 25514926.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (September 1997). "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs". Nucleic Acids Research. 25 (17): 3389–3402. doi:10.1093/nar/25.17.3389. PMC 146917. PMID 9254694.
- Yi Z, Ouyang J, Sun W, Li S, Xiao X, Zhang Q (June 2020). "Comparative exome sequencing reveals novel candidate genes for retinitis pigmentosa". EBioMedicine. 56: 102792. doi:10.1016/j.ebiom.2020.102792. PMC 7248430. PMID 32454406.