Chrysochromulina
Chrysochromulina is a genus of haptophytes. This phytoplankton is distributed globally in brackish and marine waters across approximately 60 known species.[2][3] All Chrysochromulina species are phototrophic, however some have been shown to be mixotrophic, including exhibiting phagotrophy under certain environmental conditions.[3] The cells are small, characterized by having scales, and typically observed using electron microscopy.[2][3] Some species, under certain environmental conditions have been shown to produce toxic compounds that are harmful to larger marine life including fish.[2][3][4]
| Chrysochromulina | |
|---|---|
 | |
| Chrysochromulina | |
| Scientific classification | |
| Domain: | |
| (unranked): | |
| Class: | |
| Order: | Prymnesiales |
| Family: | |
| Genus: | Chrysochromulina Lackey, 1939[1] |
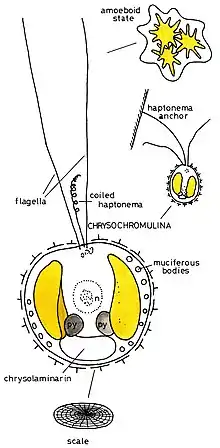
Morphology
Individuals of the genus are known to grow between 3.0 and 13.0 µm in length, with the largest being those of the Chrysochromulina polylepis species.[2] The cell surface is covered with plate-like scales, with additional layers of different scale types often overlaid.[2]
As is characteristic of all haptophytes, members of the genus Chrysochromulina possess two flagella and a unique flagella-like organelle known as the haptonema.[5] The haptonema can vary widely in length, reaching upwards of 60 µm,[2] and functions in cell attachment and feeding but differs from flagella in terms of microtubule arrangement,[5]
Ecological Significance
Chrysochromulina, as one genus of haptophytes, holds an essential role in global carbon sequestration and toxic bloom formation in world's ocean. Most haptophytes are photosynthetic micro-alga while some of them are mixotrophic.[6] Haptophytes can live in both fresh and marine water systems. This combined lifestyle makes haptophytes efficient organisms in global carbon fixation, and they occupy 30% to 50% photosynthetic biomass in the ocean.[7]
Haptophytes have an evolutionary history around 1.2 billion years long.[6] The evidence from fossils support this statement. In 2014, The draft genome sequence of Chrysochromulina tobinii has been posted by researchers from University of Washington.[6][8] C. tobinii belongs to the taxon Prymnesiales. As the first complete genome graph in this taxon, it can provide a broad understanding of haptophytes’ evolutionary history and the diversity of this clade of algae. Furthermore, it promoted the study about certain genomes and proteins which are responsible for the toxic formation and chemical release.[8]
Toxicity
Some species, such as Chrysochromulina polylepis, have been identified to produce a carbon-heavy membrane damaging toxin.[4] Research has suggested a correlation between marine nitrogen/phosphorus compositions and toxin production levels of these haptophytes; previously recorded high levels of nitrate combined with a low concentration of phosphorus led to rises in toxicity during algal blooms.[9] Further research has since determined that both low nitrogen or low phosphorus levels in the cells capable of leading to an increase in toxin production, with phosphorus proving to be slightly more influential.[4]
Despite this correlation, it unlikely that nitrogen or phosphorus are directly linked to toxin formulation, as the toxins themselves are heavily carbon-based.[4] Additionally, other growth-limiting factors such as light and salinity have also been known to increase toxicity, suggesting that the toxins where selective advantage for cell defense during times of low growth.[4] As such, studies support the idea that the metabolic responses to cellular stresses on an environmental and physiological level due to nutrient limitations are responsible for such toxin productions.[4]
Chrysochromulina blooms
Many Chrysochromulina species have been found to form algal blooms around the world.[10] Some of these blooms in the North Atlantic can produce compounds that are toxic to other marine organisms under the correct environmental conditions.[3][10][11] It is common for blooms to be formed between April and August in Scandinavian coastal waters, however the specific Chrysochromulina species present varies from year to year.[3][10]
Toxic bloom of 1988
In the late spring of 1988 the Chrysochromulina bloom that travelled from the Kattegat to the Skagerrak was made up of only one species, C. polylepis.[10][11] This particular bloom was toxic to other marine organisms including protozoa, invertebrates, and 900 tonnes of farmed fish due to the production of haemolytic compounds by C. polylepis.[10][11] C. polylepis is not typically toxic at the concentrations commonly found in the region, however certain environmental conditions such as strong stratification with a warm surface layer and low salinity following a winter featuring high amounts of nitrogen run-off increasing the N:P ratio is believed to have led to the successful C. plylepis bloom.[3][10][11] It is also thought that the production of these toxic compounds limited grazing of C. polylepis allowing for the bloom to be dominated by a single species.[10] The toxic effects seemed to reverse quickly and the food web was restored by 1993.[10][11]
Kattegat bloom in 1992
From April to May in 1992, in the southern Kattegat there was a large bloom made up of many phytoplankton species, with over 90% biomass being Chrysochromulina species.[12] The most abundant species in the bloom were C. hirta, C. spinifera, C. ericina, C. brevifilum and an undescribed species.[12] C. hirta, C. spinifera, and C. ericina are characterized as relatively small cells with long spines protruding to give the overall organisms a 25-76μm diameter which is too large for the ciliates present to engulf which is likely one reason that the bloom was so successful.[12] Another likely reason for the success of the bloom was the low presence of grazers in the bloom, about 5% of the Chrysochromulina species.[12] There was no evidence directly correlating this bloom or the species present to the production of toxins like the C. polylepis bloom in 1988.[11][12]
Major viral pathogens
Two major viruses have been found to infect Chrysochromulina: CpV-BQ1 and CeV-01B.[13][14] Freshwater samples from Lake Ontario were filtered and analyzed using transmission electron microscopy to identify the CpV-BQ1 virus. CpV-BQ1 is an icosahedral nucleocytoplasmic large DNA virus with a genome size 485kb. It is a member of the Megavirales order with characteristics of phycodnaviridae and mimivirus families. Concentrations of Chrysochromulina Lake Ontario were found to be consistent, while the CpV-BQ1 concentrations varied greatly.[13]
CeV-01B was first isolated from coastal Norwegian waters in 1998. It is an icosahedral double stranded DNA virus with a genome size of 474kb. CeV-01B belongs to a subclade of the Megaviridae family.[14]
References
- Lackey, J.B. (1939). "Notes on plankton flagellates from the Scioto River (with descriptions of new forms)". Lloydia. 2: 128–143.
- Chrétiennot-Dinet, Marie-Josèphe; Desreumaux, Nicolas; Vignes-Lebbe, Régine (2014-02-13). "An interactive key to the Chrysochromulina species (Haptophyta) described in the literature". PhytoKeys (34): 47–60. doi:10.3897/phytokeys.34.6242. PMC 3941069. PMID 24596492.
- Dahl, Einar; Bagøien, Espen; Edvardsen, Bente; Stenseth, Nils Chr. (July 2005). "The dynamics of Chrysochromulina species in the Skagerrak in relation to environmental conditions". Journal of Sea Research. 54 (1): 15–24. Bibcode:2005JSR....54...15D. doi:10.1016/j.seares.2005.02.004.
- Johansson, N.; Granéli, E. (November 1999). "Cell density, chemical composition and toxicity of Chrysochromulina polylepis (Haptophyta) in relation to different N:P supply ratios". Marine Biology. 135 (2): 209–217. doi:10.1007/s002270050618. S2CID 15796191.
- Andersen, Robert A. (2004-10-01). "Biology and systematics of heterokont and haptophyte algae". American Journal of Botany. 91 (10): 1508–1522. doi:10.3732/ajb.91.10.1508. ISSN 1537-2197. PMID 21652306.
- Hovde, Blake T.; Starkenburg, Shawn R.; Hunsperger, Heather M.; Mercer, Laina D.; Deodato, Chloe R.; Jha, Ramesh K.; Chertkov, Olga; Monnat, Raymond J.; Cattolico, Rose Ann (2014-07-17). "The mitochondrial and chloroplast genomes of the haptophyte Chrysochromulina tobin contain unique repeat structures and gene profiles". BMC Genomics. 15: 604. doi:10.1186/1471-2164-15-604. ISSN 1471-2164. PMC 4226036. PMID 25034814.
- Liu, Hui; Probert, Ian; Uitz, Julia; Claustre, Hervé; Aris-Brosou, Stéphane; Frada, Miguel; Not, Fabrice; de Vargas, Colomban (2009-08-04). "Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans". Proceedings of the National Academy of Sciences. 106 (31): 12803–12808. Bibcode:2009PNAS..10612803L. doi:10.1073/pnas.0905841106. PMC 2722306. PMID 19622724.
- Hovde, Blake T.; Deodato, Chloe R.; Hunsperger, Heather M.; Ryken, Scott A.; Yost, Will; Jha, Ramesh K.; Patterson, Johnathan; Jr, Raymond J. Monnat; Barlow, Steven B. (2015-09-23). "Genome Sequence and Transcriptome Analyses of Chrysochromulina tobin: Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae)". PLOS Genetics. 11 (9): e1005469. doi:10.1371/journal.pgen.1005469. ISSN 1553-7404. PMC 4580454. PMID 26397803.
- Skau, Lars Fredrik; Andersen, Tom; Thrane, Jan-Erik; Hessen, Dag Olav (2017-09-05). "Growth, stoichiometry and cell size; temperature and nutrient responses in haptophytes". PeerJ. 5: e3743. doi:10.7717/peerj.3743. PMC 5590550. PMID 28890852.
- Gjøsæter, Jakob; Lekve, Kyrre; Stenseth, Nils Chr; Leinaas, Hans Petter; Christie, Hartvig; Dahl, Einar; Danielssen, Didrik S.; Edvardsen, Bente; Olsgard, Frode (2000-11-22). "A long-term perspective on the Chrysochromulina bloom on the Norwegian Skagerrak coast 1988: a catastrophe or an innocent incident?". Marine Ecology Progress Series. 207: 201–218. Bibcode:2000MEPS..207..201G. doi:10.3354/meps207201. ISSN 0171-8630.
- Nielsen, Torkel; Kiørboe, Thomas; Bjørnsen, Peter (1990). "Effects of a Chrysochromulina polylepis subsurface bloom on the planktonic community" (PDF). Marine Ecology Progress Series. 62 (1/2): 21–35. Bibcode:1990MEPS...62...21N. doi:10.3354/meps062021.
- Hansen, Per; Nielsen, Torkel; Kaas, Hanne (1995). "Distribution and Growth of Protists and Mesozooplankton during a Bloom of Chrysochromulina Spp. (Prymnesiophyceae, Prymnesiales)". Phycologia. 34 (5): 409–416. doi:10.2216/i0031-8884-34-5-409.1.
- Mirza, S.F.; Staniewski, M.A.; Short, C.M.; Long, A.M.; Chaban, Y.V.; Short, S.M. (December 2015). "Isolation and characterization of a virus infecting the freshwater algae Chrysochromulina parva". Virology. 486: 105–115. doi:10.1016/j.virol.2015.09.005. PMID 26432023.
- Gallot-Lavallée, Lucie; Pagarete, António; Legendre, Matthieu; Santini, Sebastien; Sandaa, Ruth-Anne; Himmelbauer, Heinz; Ogata, Hiroyuki; Bratbak, Gunnar; Claverie, Jean-Michel (2015-12-03). "The 474-Kilobase-Pair Complete Genome Sequence of CeV-01B, a Virus Infecting Haptolina (Chrysochromulina) ericina (Prymnesiophyceae)". Genome Announcements. 3 (6). doi:10.1128/genomeA.01413-15. PMC 4669402. PMID 26634761.