Combinatorial biology
In biotechnology, combinatorial biology is the creation of a large number of compounds (usually proteins or peptides) through technologies such as phage display. Similar to combinatorial chemistry, compounds are produced by biosynthesis rather than organic chemistry. This process was developed independently by Richard A. Houghten and H. Mario Geysen in the 1980s. Combinatorial biology allows the generation and selection of the large number of ligands for high-throughput screening.[1][2]
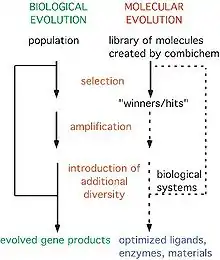
Combinatorial biology techniques generally begin with large numbers of peptides, which are generated and screened by physically linking a gene encoding a protein and a copy of said protein. This could involve the protein being fused to the M13 minor coat protein pIII, with the gene encoding this protein being held within the phage particle. Large libraries of phages with different proteins on their surfaces can then be screened through automated selection and amplification for a protein that binds tightly to a particular target.[3]
Notes
- Nill, Kimball R. (2002). Glossary of Biotechnological Terms. CRC Press. pp. 55, 56. ISBN 1-58716-122-2.
- Seethala, Ramakrishna; Fernandes, Prabhavathi B. (2001). Handbook of Drug Screening. Informa Health Care. pp. 357–383. ISBN 0-8247-0562-9.
- Pelletier J, Sidhu S (August 2001). "Mapping protein-protein interactions with combinatorial biology methods". Curr. Opin. Biotechnol. 12 (4): 340–7. doi:10.1016/S0958-1669(00)00225-1. PMID 11551461.