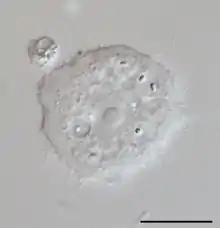
Cutosea
Cutosea (from Latin cutis 'skin') is a small group of marine amoeboid protists proposed in 2016. It is a monotypic class of Amoebozoa containing the order Squamocutida (from Latin squama 'scale', and cutis 'skin'). Cutosean organisms are characterized by a cell coat of microscales separated from the cell membrane. Three genera, Armaparvus, Sapocribrum and Squamamoeba, belong to this group, distributed in two families.
| Cutosea | |
|---|---|
| Scientific classification | |
| Domain: | Eukaryota |
| Phylum: | Amoebozoa |
| Clade: | Evosea |
| Superclass: | Cutosa Cavalier-Smith, 2016[1] |
| Class: | Cutosea Cavalier-Smith, 2016[1] |
| Order: | Squamocutida Cavalier-Smith, 2016[1] |
| Families[1] | |
| Diversity | |
| 3 species | |
Characteristics
The cells of cutosean amoebae are surrounded by a continuous thin, somewhat flexible envelope, unique in structure because it is not attached to the cytoplasmic membrane. Below this evelope, they present oval microscales surrounded by a dense matrix. The small scales are not visible under a light microscope.[2] The envelope is penetrated by one or many small pores, which allow subpseudopodia to occasionally protrude from the cell membrane,[3] for a very slow locomotion. Locomoting cells are flattened, oval or round in shape. All of their cells lack cilia or centrosomes.[1]
Taxonomy
History
Cutosea is a clade discovered through a 2016 phylogenetic study by Thomas Cavalier-Smith and his coauthors, published in the journal Molecular Phylogenetics and Evolution. It is described as a class-level rank, monotypic as it contains only one order Squamamoebida. Additionally, it is grouped under the monotypic superclass Cutosa. In traditional rank-based classifications, it is grouped within the paraphyletic Lobosa, a subphylum of Amoebozoa that also contains Tubulinea and Discosea. Cutosean amoebae present a structurally unique cellular envelope, their distinguishing feature. Their names derive from the Latin cutis, meaning 'skin', and squama, meaning 'scale', referencing this envelope.[1]
The Cutosea clade is supported by posterior molecular and morphological studies, and has been accepted as of 2019 by the International Society of Protistologists, which revises the modern cladistic classification of eukaryotes.[3] The first genera to be grouped within Cutosea were Sapocribrum and Squamamoeba, discovered in 2015 and 2013 respectively, and placed in separate families Sapocribridae and Squamamoebidae. A third genus was discovered later in 2018, Armaparvus, which was added to Squamamoebidae.[2]
Classification
Cutosea contains a total of three species, distributed in three monotypic genera:
- Family Sapocribridae Cavalier-Smith, 2016
- Sapocribrum Lahr, Grant, Molestina & Anderson, 2015[4]
- Sapocribrum chincoteaguense Lahr, Grant, Molestina & Anderson, 2015
- Sapocribrum Lahr, Grant, Molestina & Anderson, 2015[4]
- Family Squamamoebidae Cavalier-Smith, 2016
Evolution
Cutosea is a fully supported clade within Amoebozoa. It is the sister group to Conosa, which contains all the non-lobose amoebozoans: Variosea, Archamoebea and Eumycetozoa. Together, Cutosea and Conosa are the two members of the larger clade Evosea.[6] The following cladogram is based on a 2022 phylogenetic analysis:[7]
| Amorphea |
|
|||||||||||||||||||||||||||||||||||||||||||
References
- Cavalier-Smith T, Chao EE, Lewis R (June 2016). "187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution". Molecular Phylogenetics and Evolution. 99: 275–296. doi:10.1016/j.ympev.2016.03.023. PMID 27001604.
- Schuler, Gabriel A.; Brown, Matthew W. (2019). "Description of Armaparvus languidus n. gen. n. sp. Confirms Ultrastructural Unity of Cutosea (Amoebozoa, Evosea)". The Journal of Eukaryotic Microbiology. 66: 158–166. doi:10.1111/jeu.12640.
- Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. PMC 6492006. PMID 30257078.
- Lahr, Daniel J. G.; Grant, Jessica; Molestina, Robert; Katz, Laura A.; Anderson, O. Roger. "Sapocribrum chincoteaguense n. gen. n. sp.: A Small, Scale-bearing Amoebozoan with Flabellinid Affinities". The Journal of Eukaryotic Microbiology. 62: 444–453. doi:10.1111/jeu.12199.
- Kudryavtsev, Alexander; Pawlowski, Jan (2013). "Squamamoeba japonica n. g. n sp. (Amoebozoa): a deep-sea amoeba from the Sea of Japan with a novel cell coat structure". Protist. 164 (1): 13–23. doi:10.1016/j.protis.2012.07.003.
- Kang, Seungho; Tice, Alexander K; Spiegel, Frederick W; Silberman, Jeffrey D; Pánek, Tomáš; Čepička, Ivan; Kostka, Martin; Kosakyan, Anush; Alcântara, Daniel M C; Roger, Andrew J; Shadwick, Lora L; Smirnov, Alexey; Kudryavtsev, Alexander; Lahr, Daniel J G; Brown, Matthew W (September 2017). "Between a Pod and a Hard Test: The Deep Evolution of Amoebae". Molecular Biology and Evolution. 34 (9): 2258–2270. doi:10.1093/molbev/msx162. PMC 5850466. PMID 28505375.
- Tekle YI, Wang F, Wood FC, Anderson OR, Smirnov A (2022). "New insights on the evolutionary relationships between the major lineages of Amoebozoa". Sci Rep. 12 (11173): 11173. Bibcode:2022NatSR..1211173T. doi:10.1038/s41598-022-15372-7. PMC 9249873. PMID 35778543. S2CID 247231712.
External links
- Cutosea at UniEuk Taxonomy App.