D-Ribose pyranase
D-Ribose pyranase is an enzyme that catalyzes the interconvesion of β-D-ribopyranose and β-D-ribofuranose.[1] This enzyme is an isomerase that has only been found in bacteria and viruses. It has two known functions of helping transport ribose into cells and producing β-D-ribofuranose, which can later be used to make ribose 5-phosphate for the pentose phosphate pathway (PPP). D-Ribose pyranase does not have a defined crystal structure but there are two different proposed structures. The active site of D-ribose pyranase is high in histidine residues along with a few other key binding sites.
EC number
The Enzyme Commission (EC) number is a way to classify enzymes in a numerical fashion. It is based on the type of chemical reactions the enzyme catalyzes. For D-ribose pyranase, the EC number is 5.4.99.62. The first number is the general type of reaction catalyzed by the enzyme. The second and third numbers are related to specific details about the actual reaction the enzyme catalyzes. More specifically, the second number is the subclass and the third number is the sub-subclass. The fourth number is a serial identifier. Here is a breakdown of the EC number for D-ribose pyranase:[2]
- 5 Isomerases
- 5.4 Intramolecular transferases
- 5.4.99 Transferring other groups
- 5.4.99.62 D-Ribose Pyranase
- 5.4.99 Transferring other groups
- 5.4 Intramolecular transferases
To understand the specifics of D-ribose pyranase, a description of each of the EC parts will now be provided. An isomerase is an enzyme that catalyzes a reaction in which a molecular rearrangement takes place. Intermolecular transferases are enzymes that catalyze the transfer of acyl-, phospho-, amino-, and other groups from one spot to another on a molecule. Transferring other groups deals with the specific area on the molecule the enzyme is manipulating. Finally, the end result of the EC number is the enzyme of D-Ribose Pyranase[2].
Enzyme reaction pathway

The main reaction that D-ribose pyranase catalyzes is the reversible reaction:[1]
- β-D-ribopyranose ⇌ β-D-ribofuranose
The main event in this reaction is that a six-membered ring (pyranose) gets converted into a five-membered ring (furanose). This is why pyranoses and furanoses are considered structural isomers of each other. This means they have the same atoms but are connected in a different way. In the reaction, β-D-ribopyranose gets protonated to make a cationic intermediate. After shifting electrons from an electron donating group, the pyranose ring opens. Next, a nucleophilic addition reaction occurs. Lastly, a deprotonation takes place, leaving the newly formed β-D-ribofuranose in a neutral, stable state[4]. It can be noted that both beta-D-ribofuranose and beta-D-ribopyranose both have the chemical formula of C5H10O5. This shows how they are isomers of each other.[1]
The enzyme can also catalyze the reaction:[1]
- β-allopyranose ⇌ β-allofuranose
Organisms in which D-ribose pyranase is found
D-Ribose pyranase is found in a variety of organisms ranging from viruses to bacteria. This enzyme has been mostly studied in Escherichia coli (E. Coli), Bacillus subtilis (B. subtilis), and Staphylococcus aureus (S. aureus).[5] D-Ribose pyranase is part of an RBS operon that is responsible for transporting ribose into the cell. The RBS operon uses active transport to get ribose into the cell. All of the organisms listed previously have this system in which can facilitate the conversion of the sugar ribose into a specific form; either the furanose or pyranose form. Ribose enters the cell in the pyranose form, so its crucial that D-ribose pyranase is present in the cell to convert it to the furanose form. Additionally, these bacteria need to convert the pyranose to the furanose form since the furanose form is utilized in the pentose phosphate pathway (PPP).[2] The PPP is described in the next section.
Function in the cell
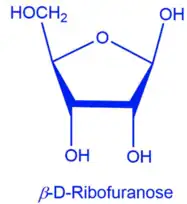
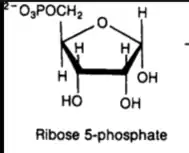
Ribose can either be a five membered ring (furanose) or a six membered ring (pyranose). In terms of cells, the furanose form is more useful since it can be used in further reactions in the cell. For most cells, ribose is transported into the cell in the pyranose form. With this said, D-Ribose Pyranase needs to be present to convert the pyranose form into the furanose form. Beta-D-ribofuranose can then be converted to ribose-5-phosphate. In the images on the right, both structures have five carbons but differ in the amount of hydrogens by one, oxygens by three, and phosphorus by one. The key difference is the group on carbon four. From here, ribose 5-phosphate can enter into the pentose phosphate pathway (PPP)[7]. The PPP is a major system in metabolism that can make precursor metabolites, it has reducing power, and can produce metabolic energy. More specifically, the PPP is able to produce NADPH. NADPH is an electron donor that is able to supply reducing abilities in anabolic reactions and helps balance out redox reactions. NADPH is able to aid in the production of things like lipids and amino acids. Additionally, ribose 5-phosphate is able to help make nucleic acids[8]. Therefore, the role of D-ribose pyranase is early on in the cascade of the PPP. Without D-ribose pyranase, production of many vital cell components would not be produced in a timely manner.
Known crystal structure

There are many different proposed crystal structures of D-ribose pyranase, depending on the methods the structure was researched and what organism it was researched in. In a study done by Wang et. al, it was thought that the structure of D-Ribose Pyranase in S. aureus is a dimeric structure based off of a sitting-drop vapour-diffusion test[9].
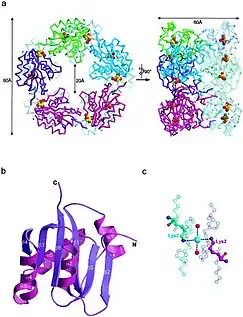
The proposed crystal structure of D-ribose pyranaseby Kim et. al. using B. subtilis is a protein in a homodecameric assembly with two major units. The asymmetric portion of the protein crystal structure is a pentameric ring structure. This pentameric structure is connected to another pentameric ring. There are very tight interactions between these two units. It has five glycerol molecules, two chloride ions, and 369 water molecules. It was originally believed that D-ribose pyranase was an octamer in solution in terms of being investigated in E. Coli. This new study by Kim et. al. showed that the structure of the asymmetric portion's subunits are an alpha beta fold. It has a central six-stranded beta sheet in between three alpha helices on one side, and both an alpha helix and a beta strand loop on the other side. There is an area in the enzyme structure that forms a sort of "cage." It has a lysine residue and water molecules which are brought together to form the cage due to a chloride ion binding. This cave is a crucial part of the stability of the protein structure. On the image on the right, the top two pictures are decameric structure of D-Ribose Pyranase[10].
Known active sites
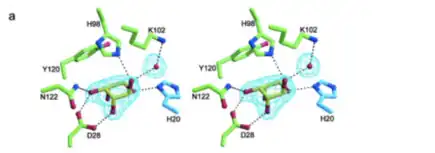
The active site of D-ribose pyranase is at position 20 and is a proton donor. The amino acid at this location is a histidine. This amino acid has two nitrogen’s that are able to act as proton donors. There are other binding sites at positions 28, 98, 102,106,120,122, and 128-130 in the amino acid chain. Position 28 is an aspartic acid residue, position 98 is a histidine, position 102 is a lysine, position 106 is a histidine residue, position 120 is a tyrosine, position 122 is an asparagine, and positions 128-130 are tryptophan, alanine, and asparagine[11]. It has been found that Tyr-120 is positioned very close to the ribose sugar when it is bound to the D-Ribose Pyranase since it provides a hydrophobic environment. His-20 and His-106 are thought of as being highly conserved. These positions are similar to that of the enzyme of FucU, which is another enzyme family that is similar to that of D-Ribose Pyranase[10]. These two positions are crucial for the binding of ribose. In an experiment by Ryu et. al, it was found that when these two positions were mutated to alanine instead of histidine, the catalytic activity of D-Ribose Pyranase was drastically reduced[12].
References
- "EC 5.4.99.62". iubmb.qmul.ac.uk. Retrieved 2023-10-23.
- Bank, RCSB Protein Data. "RCSB PDB - 3P12: Crystal Structure of D-ribose Pyranase Sa240". www.rcsb.org. Retrieved 2023-10-23.
- "Exam 2 Answer Key". web.pdx.edu. Retrieved 2023-10-23.
- "Information on EC 5.4.99.62 - D-ribose pyranase - BRENDA Enzyme Database". www.brenda-enzymes.org. Retrieved 2023-10-23.
- "D-Ribose Pyranase". EcoCyc.
- https://www.sciencedirect.com/topics/medicine-and-dentistry/ribose-5-phosphate
- McLeod, Anette; Zagorec, Monique; Champomier-Vergès, Marie-Christine; Naterstad, Kristine; Axelsson, Lars (April 2010). "Primary metabolism in Lactobacillus sakei food isolates by proteomic analysis". BMC Microbiology. 10 (1): 1–10. doi:10.1186/1471-2180-10-120. ISSN 1471-2180.
- "NADPH: Definition, Structure & Function". Study.com.
- Wang, Ling; Wu, Minhao; Zang, Jianye (2011-05-01). "Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus". Journal of Structural Biology. 174 (2): 413–419. doi:10.1016/j.jsb.2011.01.007. ISSN 1047-8477.
- Kim, Min-Sung; Shin, Joon; Lee, Weontae; Lee, Heung-Soo; Oh, Byung-Ha (2003-07-25). "Crystal Structures of RbsD Leading to the Identification of Cytoplasmic Sugar-binding Proteins with a Novel Folding Architecture*". Journal of Biological Chemistry. 278 (30): 28173–28180. doi:10.1074/jbc.M304523200. ISSN 0021-9258.
- "UniProt". www.uniprot.org. Retrieved 2023-10-23.
- Ryu, Kyoung-Seok (June 2024). "NMR Application Probes a Novel and Ubiquitous Family of Enzymes That Alter Monosaccharide Configuration". Journal of Biological Chemistry.