dNaM
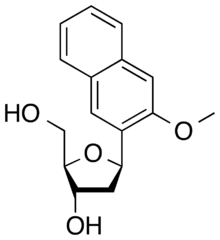
dNaM is an artificial nucleobase containing a 3-methoxy-2-naphthyl group instead of a natural base.
 | |
| Names | |
|---|---|
| IUPAC name
(1R)-1,4-Anhydro-2-deoxy-1-(3-methoxynaphthalen-2-yl)-D-erythro-pentitol | |
| Systematic IUPAC name
(2R,3S,5R)-2-(Hydroxymethyl)-5-(3-methoxynaphthalen-2-yl)oxolan-3-ol | |
| Identifiers | |
3D model (JSmol) |
|
| ChemSpider | |
PubChem CID |
|
| |
| |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
When it was originally successfully introduced into DNA for replication in an E. coli semi-synthetic organism, it was paired up with d5SICS. For short it is called X whilst the d5SICS being called Y.[1] d5SICS was replaced by dTPT3 in revised versions due to its improved ability to replicate in a wider range of sequence contexts.[2] X pairs with Y using hydrophobic and packing interactions instead of hydrogen bonding, which occurs in natural base pairs. Inside the semi-synthetic organism, methyl directed mismatch repair pathway (MMR) actually fixes unnatural-natural mispairs, whereas recombinational repair actually cuts out the unnatural.[3] The E. coli semi-synthetic organism managed to hold onto the new base for an extended time both while on a plasmid as well as when stored in the chromosome.[4][5] In free DNA, rings of d5SICS and dNaM are placed in parallel planes instead of the same plane, but when inside of a DNA polymerase, they pair using an edge-to-edge conformation.[6] dNaM and dTPT3 can also template transcription of mRNAs and tRNAs by T7 RNA polymerase that have the ability to produce decode at the E. coli ribosome to produce proteins with unnatural amino acids, expanding the genetic code.[7]
References
- Sarah Caplan (29 November 2017). "Cells with DNA made in lab lead to 'Holy Grail' of synthetic biology". The Washington Post.
- Zhang, Yorke; Lamb, Brian M.; Feldman, Aaron W.; Zhou, Anne Xiaozhou; Lavergne, Thomas; Li, Lingjun; Romesberg, Floyd E. (2017-01-23). "A semisynthetic organism engineered for the stable expansion of the genetic alphabet". Proceedings of the National Academy of Sciences. 114 (6): 1317–1322. doi:10.1073/pnas.1616443114. ISSN 0027-8424. PMC 5307467. PMID 28115716.
- Ledbetter, Michael P.; Karadeema, Rebekah J.; Romesberg, Floyd E. (2018-01-17). "Reprograming the Replisome of a Semisynthetic Organism for the Expansion of the Genetic Alphabet". Journal of the American Chemical Society. 140 (2): 758–765. doi:10.1021/jacs.7b11488. ISSN 0002-7863. PMC 5793209. PMID 29309130.
- "Bacterium survives unnatural DNA transplant". Rsc.org. Retrieved July 29, 2015.
- Malyshev, Denis A.; Dhami, Kirandeep; Quach, Henry T.; Lavergne, Thomas; Ordoukhanian, Phillip; Torkamani, Ali; Romesberg, Floyd E. (2012). "Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet". Proceedings of the National Academy of Sciences. 109 (30): 12005–12010. Bibcode:2012PNAS..10912005M. doi:10.1073/pnas.1205176109. PMC 3409741. PMID 22773812. S2CID 26653524.
- Betz, Karin; et al. (2013). "Structural Insights into DNA Replication Without Hydrogen-Bonds". J Am Chem Soc. 135 (49): 18637–43. doi:10.1021/ja409609j. PMC 3982147. PMID 24283923.
- Zhang, Yorke; Ptacin, Jerod L.; Fischer, Emil C.; Aerni, Hans R.; Caffaro, Carolina E.; San Jose, Kristine; Feldman, Aaron W.; Turner, Court R.; Romesberg, Floyd E. (November 2017). "A semi-synthetic organism that stores and retrieves increased genetic information". Nature. 551 (7682): 644–647. doi:10.1038/nature24659. ISSN 1476-4687. PMC 5796663. PMID 29189780.