R.EcoRII
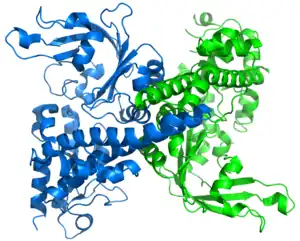
Restriction endonuclease (REase) EcoRII (pronounced "eco R two") is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.[1]
Mode of action
EcoRII is a bacterial Type IIE[2] REase that interacts with two[3] or three[4] copies of the pseudopalindromic DNA recognition sequence 5'-CCWGG-3' (W = A or T), one being the actual target of cleavage, the other(s) serving as the allosteric activator(s). EcoRII cuts the target DNA sequence CCWGG, generating sticky ends.[5]
Cut diagram
| Recognition site | Cut results |
5' NNCCWGGNN 3' NNGGWCCNN |
5' NN CCWGGNN 3' NNGGWCC NN |
Structure
| Restriction endonuclease EcoRII, N-terminal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
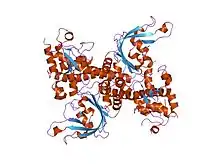 crystal structure of restriction endonuclease ecorii mutant r88a | |||||||||
| Identifiers | |||||||||
| Symbol | EcoRII-N | ||||||||
| Pfam | PF09217 | ||||||||
| Pfam clan | CL0405 | ||||||||
| InterPro | IPR015300 | ||||||||
| SCOP2 | 1na6 / SCOPe / SUPFAM | ||||||||
| |||||||||
| EcoRII C terminal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of restriction endonuclease ecorii mutant r88a | |||||||||
| Identifiers | |||||||||
| Symbol | EcoRII-C | ||||||||
| Pfam | PF09019 | ||||||||
| Pfam clan | CL0236 | ||||||||
| InterPro | IPR015109 | ||||||||
| |||||||||
The apo crystal structure of EcoRII mutant R88A (PDB: 1NA6)[6] has been solved at 2.1 Å resolution. The EcoRII monomer has two domains, N-terminal and C-terminal, linked through a hinge loop.
Effector-binding domain
The N-terminal effector-binding domain has an archetypal DNA-binding pseudobarrel fold (SCOP 101936) with a prominent cleft. Structural superposition showed it is evolutionarily related to:
- B3 DNA binding domain (SCOP 117343) from the transcription factors in higher plants (PDB: 1WID)[7]
- C-terminal domain of restriction endonuclease BfiI[8] (PDB: 2C1L)[9]
Catalytic domain
The C-terminal catalytic domain has a typical[10] restriction endonuclease-like fold (SCOP 52979) and belongs to the large (more than 30 members) restriction endonuclease superfamily (SCOP 52980).
Autoinhibition/activation mechanism
Structure-based sequence alignment and site-directed mutagenesis identified the putative PD..D/EXK active sites of the EcoRII catalytic domain dimer that in apo structure are spatially blocked by the N-terminal domains.[6]
See also
- EcoRI, another nuclease enzyme from Escherichia coli.
- EcoRV, another nuclease enzyme from Escherichia coli.
- B3 DNA binding domain from higher plants is evolutionary related to EcoRII
- FokI, another nuclease enzyme from Flavobacterium okeanokoites
External links
- EcoRII in Restriction Enzyme Database REBASE
References
- Richard J. Roberts. "EcoRII". REBASE - The Restriction Enzyme Database. Retrieved 2008-03-23.
- Roberts RJ, Belfort M, Bestor T, et al. (2003). "A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes". Nucleic Acids Res. 31 (7): 1805–12. doi:10.1093/nar/gkg274. PMC 152790. PMID 12654995. PDF
- Mücke M, Lurz R, Mackeldanz P, Behlke J, Krüger DH, Reuter M (2000). "Imaging DNA loops induced by restriction endonuclease EcoRII. A single amino acid substitution uncouples target recognition from cooperative DNA interaction and cleavage". J. Biol. Chem. 275 (39): 30631–7. doi:10.1074/jbc.M003904200. PMID 10903314.PDF
- Shlyakhtenko LS, Gilmore J, Portillo A, Tamulaitis G, Siksnys V, Lyubchenko YL (2007). "Direct visualization of the EcoRII-DNA triple synaptic complex by atomic force microscopy". Biochemistry. 46 (39): 11128–36. doi:10.1021/bi701123u. PMID 17845057. S2CID 27800123.
- Griffiths, Anthony J. F. (1999). An Introduction to genetic analysis. San Francisco: W.H. Freeman. ISBN 978-0-7167-3520-5.
- Zhou XE, Wang Y, Reuter M, Mücke M, Krüger DH, Meehan EJ, Chen L (2004). "Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold". J. Mol. Biol. 335 (1): 307–19. doi:10.1016/j.jmb.2003.10.030. PMID 14659759.
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Hayami N, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S (2004). "Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1". Plant Cell. 16 (12): 3448–59. doi:10.1105/tpc.104.026112. PMC 535885. PMID 15548737.PDF
- Richard J. Roberts. "BfiI". REBASE - The Restriction Enzyme Database. Retrieved 2008-03-23.
- Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V (2005). "Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease" (PDF). Proc. Natl. Acad. Sci. U.S.A. 102 (44): 15797–802. Bibcode:2005PNAS..10215797G. doi:10.1073/pnas.0507949102. PMC 1266039. PMID 16247004. PDF
- Niv MY, Ripoll DR, Vila JA, Liwo A, Vanamee ES, Aggarwal AK, Weinstein H, Scheraga HA (2007). "Topology of Type II REases revisited; structural classes and the common conserved core". Nucleic Acids Research. 35 (7): 2227–37. doi:10.1093/nar/gkm045. PMC 1874628. PMID 17369272.