FAM221A
Family with sequence similarity 221 member A is a protein in humans that is encoded by the FAM221A gene. FAM221A is a gene that is not yet well understood by the scientific community. However, it appears that this gene may have a role in Parkinson's disease and prostate cancer.
| FAM221A | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | FAM221A, C7orf46, family with sequence similarity 221 member A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 2442161 HomoloGene: 18214 GeneCards: FAM221A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
Location and Aliases
FAM221A is located on Chromosome 7. Its exact location is 7p15.3.[5] It has one alias, which is C7orf46.[6]
Expression
FAM221A has higher levels of expression in the liver, brain, fetal brain, thyroid and colon, but FAM221A has the highest level of expression in the spinal cord, pancreas and retina.[7]
The promoter region of FAM221A is 1222 base pairs long. This was found using ElDorado at Genomatix.[8]
Protein
Protein Analysis
The molecular weight of FAM221A is 33.1 kDa,[9] and the isoelectric point is 6.01.[10] Relative to other proteins in humans, FAM221A has a lower level of asparagine.[9]
Post-Translational Modifications
Post-translational modifications of FAM221A include phosphorylation sites, glycosylation sites and sulfation sites. These have been conserved in mammals other than Homo sapiens, including the macaque, whale, finch and sometimes alligator. These sites were predicted using NetPhos 3.1,[11] YinOYang 1.2[12] and The Sulfinator.[13]
Secondary Structure
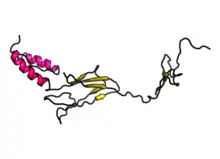
Key structures predicted in FAM221A are random coils and alpha helices, with 71% of the protein being random coils and 21% being helices. Extended strands were also found with 7% of the protein being these. Secondary structure was predicted using RaptorX,[14] and a diagram of the predicted secondary structure is included below.
Homology/evolution
Paralogs
There exists one paralog for FAM221A: FAM221B. This diverged from FAM221A approximately 1781 million years ago.
Orthologs
Orthologs have been found in mammals, birds, reptiles and fish. FAM221A has also been conserved in invertebrates, but the similarity levels decrease at a faster rate. Orthologs were discovered using BLAST [15] and BLAT.[16] While these are not the only orthologs that exist for FAM221A, a table of 20 orthologs is provided below. The ortholog with no accession number was created using BLAT.
| Species | Common Name | Divergence (mya) | Accession Number | Length (aa) | % Identity | % Similarity |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | 0 | NP_954587.2 | 298 | 100 | 100 |
| Macaca nemestrina | Southern pig-tailed macaque | 28.1 | XP_011729478.1 | 298 | 96 | 96 |
| Condylura cristata | Star-nosed mole | 94 | XP_004677186.2 | 284 | 90 | 94 |
| Cervus elaphus hippelaphus | Central European red deer | 94 | OWK06795.1 | 289 | 90 | 93 |
| Delphinapterus leucas | Beluga whale | 94 | XP_022440764.1 | 298 | 90 | 92 |
| Alligator mississippiensis | American alligator | 320 | KYO26809.1 | 366 | 78 | 86 |
| Phalacrocorax carbo | Great cormorant | 320 | KFW96932.1 | 258 | 77 | 87 |
| Lonchura striata domestica | Society finch | 320 | XP_021393915.1 | 298 | 76 | 85 |
| Pelodiscus sinensis | Chinese softshell turtle | 320 | XP_014436679.1 | 236 | 76 | 85 |
| Gallus Gallus | Red junglefowl | 320 | XP_418719.1 | 296 | 75 | 84 |
| Crocodylus porosus | Saltwater crocodile | 320 | XP_019390202.1 | 236 | 75 | 84 |
| Amphiprion ocellaris | Ocellaris clownfish | 432 | XP_023141881.1 | 248 | 63 | 75 |
| Salvelinus alpinus | Arctic char | 432 | XP_023832019.1 | 372 | 59 | 71 |
| Esox lucius | Northern pike | 432 | XP_010891304.1 | 332 | 55 | 69 |
| Ciona intestinalis | Vase tunicate | 678 | N/A | 212 | 77 | 87 |
| Stylophora pistillata | Stylophora pistillata | 685 | XP_022787363.1 | 344 | 58 | 73 |
| Schistosoma haematobium | Uniary blood fluke | 692 | XP_012794504.1 | 241 | 45 | 61 |
| Crassostrea virginica | Eastern oyster | 794 | XP_022337450.1 | 324 | 59 | 72 |
| Mizuhopecten yessoensis | Patinopecten yessoensis | 794 | XP_021377417.1 | 326 | 55 | 70 |
| Phytophthora nicotianae | Black shank | 1781 | KUF80258.1 | 297 | 34 | 48 |
| Chrysochromulina sp. CCMP291 | Chrysochromulina tobin | 1781 | KOO33212.1 | 280 | 28 | 42 |
Divergence of FAM221A
To understand the times when FAM221A diverged from different species, a graph was created. This compares the evolutionary history of FAM221A to Fibrinogen, which evolves quickly, and Cytochrome C, which evolves slowly. As seen in the graph, FAM221A diverges from other species at a moderate pace.

Clinical significance
FAM221A has a relatively high amount of expression in the brain[17] and has been seen to have an association with neurodegenerative disorders such as Parkinson's disease[17] and Alzheimer's disease.[18] FAM221A has also been seen to have a higher level of expression in those who have prostate cancer versus healthy individuals.[19] Furthermore, FAM221A has also been expressed in those with colorectal tumors.[20]
Interacting Proteins
Three interacting proteins were found, which are SNX2, SNX5 and SNX6.
SNX2 and SNX6 share the same function, which is being involved in the stages of intracellular trafficking. SNX5 facilitates cargo retrieval from endosomes to the trans-golgi network.
References
- GRCh38: Ensembl release 89: ENSG00000188732 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000047115 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: Family with sequence similarity 221 member A". Retrieved 2016-07-20.
- Database, GeneCards Human Gene. "FAM221A Gene - GeneCards - F221A Protein - F221A Antibody". www.genecards.org.
- "GDS3113 / 125374". www.ncbi.nlm.nih.gov.
- "Genomatix". www.genomatix.de.
- "SAPS < Sequence Statistics < EMBL-EBI".
- "Calculation of Protein Isoelectric Point".
- "NetPhos 3.1 Server". www.cbs.dtu.dk.
- "YinOYang 1.2 Server". www.cbs.dtu.dk.
- "ExPASy - Sulfinator tool". web.expasy.org.
- http://raptorx.uchicago.edu/.
{{cite web}}: Missing or empty|title=(help) - "NCBI BLAST".
- "UCSC BLAT".
- Mariani E, Frabetti F, Tarozzi A, Pelleri MC, Pizzetti F, Casadei R (2016). "Meta-Analysis of Parkinson's Disease Transcriptome Data Using TRAM Software: Whole Substantia Nigra Tissue and Single Dopamine Neuron Differential Gene Expression". PLOS ONE. 11 (9): e0161567. Bibcode:2016PLoSO..1161567M. doi:10.1371/journal.pone.0161567. PMC 5017670. PMID 27611585.
- Thonberg H, Chiang HH, Lilius L, Forsell C, Lindström AK, Johansson C, Björkström J, Thordardottir S, Sleegers K, Van Broeckhoven C, Rönnbäck A, Graff C (June 2017). "Identification and description of three families with familial Alzheimer disease that segregate variants in the SORL1 gene". Acta Neuropathologica Communications. 5 (1): 43. doi:10.1186/s40478-017-0441-9. PMC 5465543. PMID 28595629.
- Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, Sanda MG (September 2009). "Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer". Clinical Cancer Research. 15 (18): 5794–802. doi:10.1158/1078-0432.CCR-09-0911. PMC 5573151. PMID 19737960.
- Khamas A, Ishikawa T, Shimokawa K, Mogushi K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H, Sugihara K (2012). "Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2'-deoxycytidine, microarray and gene expression profile". Cancer Genomics & Proteomics. 9 (2): 67–75. PMID 22399497.