Fimbrial usher protein
The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of some fimbriae (or pili) requires a two-component assembly and transport system which is composed of a periplasmic chaperone and a pore-forming outer membrane protein which has been termed a molecular 'usher'; this is the chaperone-usher pathway.[2][3][4]
| Fimbrial Usher protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
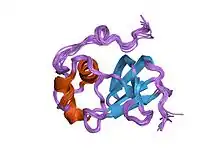 Structure of the type 1 pilus assembly platform FimD(25-139).[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Usher | ||||||||
| Pfam | PF00577 | ||||||||
| InterPro | IPR000015 | ||||||||
| PROSITE | PDOC00886 | ||||||||
| TCDB | 1.B.11 | ||||||||
| OPM superfamily | 187 | ||||||||
| OPM protein | 4j3o | ||||||||
| |||||||||
The usher protein has a molecular weight ranging from 86 to 100 kDa and is composed of a membrane-spanning 24-stranded beta barrel domain, reminiscent of porins, and of four periplasmic soluble domains: an N-terminal one of about 120 residues (NTD),[1] a 'middle' domain of about 80 residues[5] located as a soluble insertion within the beta barrel region of the sequence (plug domain) and two IG-like domains (each about 80 residues long) at the C-terminus (CTD1 and CTD2).[6] Although the degree of sequence similarity of these proteins is not very high they share a number of characteristics. One of these is the presence of two pairs of disulfide bond-forming cysteines, the first one located in the NTD and the second in CTD2. The best conserved region of the sequence corresponds to the plug domain.
References
- Nishiyama M, Horst R, Eidam O, et al. (June 2005). "Structural basis of chaperone–subunit complex recognition by the type 1 pilus assembly platform FimD". EMBO J. 24 (12): 2075–86. doi:10.1038/sj.emboj.7600693. PMC 1150887. PMID 15920478.
- Hultgren SJ, Jacob-Dubuisson F, Striker R (1994). "Chaperone-assisted self-assembly of pili independent of cellular energy". J. Biol. Chem. 269 (17): 12447–12455. PMID 7909802.
- Schifferli DM, Alrutz MA (1994). "Permissive linker insertion sites in the outer membrane protein of 987P fimbriae of Escherichia coli". J. Bacteriol. 176 (4): 1099–1110. doi:10.1128/JB.176.4.1099-1110.1994. PMC 205162. PMID 7906265.
- Saier Jr MH, Van Rosmalen M (1993). "Structural and evolutionary relationships between two families of bacterial extracytoplasmic chaperone proteins which function cooperatively in fimbrial assembly". Res. Microbiol. 144 (7): 507–527. doi:10.1016/0923-2508(93)90001-I. PMID 7906046.
- Capitani G, Eidam O, Grütter MG (2006). "Evidence for a novel domain of bacterial outer membrane ushers". Proteins. 65 (4): 816–23. doi:10.1002/prot.21147. PMID 17066380. S2CID 28766740.
- Phan G, Remaut H, Wang T, Allen WJ, Pirker KF, Lebedev A, et al. (2011). "Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate". Nature. 474 (7349): 49–53. doi:10.1038/nature10109. PMC 3162478. PMID 21637253.