Fixed allele
In population genetics, a fixed allele is an allele that is the only variant that exists for that gene in a population. A fixed allele is homozygous for all members of the population.[1] The process by which alleles become fixed is called fixation.

A population of a hypothetical species can be conceived to exemplify the concept of fixed alleles. If an allele is fixed in the population, then all organisms can have only that allele for the gene in question. Suppose that genotype corresponds directly to the phenotype of body color, then all organisms of the population would exhibit the same body color.
An allele in a population being fixed necessarily entails the phenotypic traits corresponding to that allele to be identical for all organisms in the population (if those genotypes correspond directly to a certain phenotype), as it follows logically from the definition of relevant concepts. However, identical phenotypic traits exhibited in a population does not necessarily entail the allele(s) corresponding to those traits to be fixed, as exemplified by the case of genetic dominance being apposite in a species' population. [2]
Low genetic diversity is accompanied by allele fixation, which can potentially lead to lower adaptibility to changing environmental conditions for a population as a whole. For example, often having certain alleles make an organism more susceptible to a disease than having other alleles; if an allele highly susceptible to a disease with a prevalent cause is fixed in a population, most organisms of the population might be affected. Hence, generally, populations exhibiting a significant range of fixed alleles are often at risk for extinction. [3][4]
Fixed alleles were first defined by Motoo Kimura in 1962.[5] Kimura discussed how fixed alleles could arise within populations and was the first to generalize the topic. He credits the works of Haldane in 1927[6] and Fisher in 1922[7] as being important in providing foundational information that allowed him to come to his conclusion.
The evolution of fixed alleles in populations
While there are many possibilities for how a fixed allele can develop, often multiple factors come into play simultaneously and guide the process, consequently determining the end result.
The two key driving forces behind fixation are natural selection and genetic drift. Natural selection was postulated by Charles Darwin and encompasses many processes that lead to the differential survival of organisms due to genetic or phenotypic differences. Genetic drift is the process by which allele frequencies fluctuate within populations. Natural selection and genetic drift propel evolution forward, and through evolution, alleles can become fixed.[8][9]
Processes of natural selection such as sexual, convergent, divergent, or stabilizing selection pave the way for allele fixation. One way some of these natural selection processes cause fixation is through one specific genotype or phenotype being favored, which leads to the convergence of the variability until one allele becomes fixed. Natural selection can work the other way, where two alleles become fixed through two specific genotypes or phenotypes being favored, leading to divergence within the population until the populations become so separate that they are now two species each with their own fixed allele.
Selective pressures can favor certain genotypes or phenotypes. A commonly known example of this is the process of antibiotic resistance within bacterial populations. As antibiotics are used to kill bacteria, a small number of them with favorable mutations can survive and repopulate in an environment that is now free of competition. The allele for antibiotic resistance then becomes a fixed allele within the surviving and future populations. This is an example of the bottleneck effect. A bottleneck occurs when a population is put under strong selective pressure, and only certain individuals survive. These surviving individuals have a decreased number of alleles present within their population than were present in the initial population, however, these remaining alleles are the only ones left in future populations assuming no mutation or migration. This bottleneck effect can also be seen in natural disasters, as shown in the rabbit example above.[10]
Similar to the bottleneck effect, the founder's effect can also cause allele fixation. The founder effect occurs when a small founding population is moved to a new area and propagates the future population. This can be seen in the Alces alces moose population in Newfoundland, Canada. Moose are not native to Newfoundland, and in 1878 and 1904 six total moose were introduced to the island. The six founding moose propagated the current population of an estimated 4000-6000 moose. This has had dramatic effects on the offspring of the founding moose and has led to a great decrease in genetic variability within the Newfoundland moose population as compared to the mainland population.[11][12]
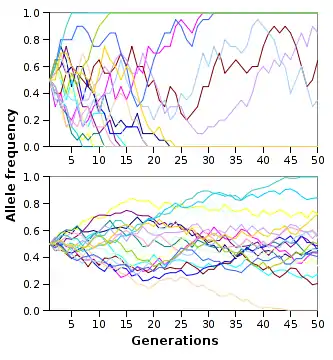
Other random processes such as genetic drift can lead to fixation. Through these random processes, some random individuals or alleles are removed from the population. These random fluctuations within the allele frequencies can lead to the fixation or loss of certain alleles within a population. To the right is an image that shows through successive generations; the allele frequencies fluctuate randomly within a population. The smaller the population size, the faster fixation or loss of alleles will occur. However, all populations are driven to allele fixation and it is inevitable; it just takes varying amounts of time for this to occur due to population size.
Some other causes of allele fixation are inbreeding, as this decreases the genetic variability of the population and therefore decreases the effective population size.[11][13] This allows genetic drift to cause fixation faster than anticipated.
Isolation can also cause fixation, as it prevents the influx of new variable alleles into the population. This can often be seen on island populations, where the populations have a limited set of alleles. The only variability that can be added to these populations is through mutations.[11][12]
Examples
One example of a fixed allele is the DGAT-1 exon 8 in Anatolian buffalo. This is a non-conservative mutation in the DGAT-1 allele, which produces a protein with a lysine at position 232 instead of an alanine. This mutation produces a protein different from the wild type of protein. This mutation in cattle affects milk production. Investigation into three water buffalo populations revealed four different haplotypes each having a single nucleotide polymorphism (SNP), however, all of these SNPs were conservative mutations, causing no change in protein production. All populations of Anatolian buffalo studied had the non-conservative lysine mutation at 232, leading to the conclusion that this DGAT-1 allele mutation is fixed within the populations.[14]
The Parnassius apollo butterfly is classified as a threatened species, having many disjointed populations in the Western Palaearctic region. The population in the Mosel Valley of Germany has been genetically characterized and had been shown to have six long-term monomorphic microsatellites. Six microsatellites were examined by looking at the current population in 2008 as well as museum samples from 1895 to 1989. One of the microsatellite alleles examined has become fixed within the population before1895. For the current population, all six microsatellites as well as all sixteen alloenzymes analyzed were fixed.[15]
Fixed alleles can often be deleterious to populations, especially when there is small population size and low genetic variability. For example, the California Channel Island Fox (Urocyon littoralis) has the most monomorphic population ever reported for a sexually reproducing animal.[15] During the 1990s the Island Fox experienced disastrous population decline, leading to near extinction.[3] This population decline was caused in part by the canine distemper virus. The foxes were susceptible to this virus, and many were killed due to their genetic similarity. The introduction of a predator, the golden eagle, was also attributed to this population decline. With current conservation efforts, the population is in recovery.[4]
See also
References
- "fixed allele definition". www.biochem.northwestern.edu.
- "Genotype & Phenotype". biomed.brown.edu. Retrieved 2023-05-25.
- Coonan, Timothy; Schwemma, Catherin; Roemerb, Gary; Garcelonc, David; Munsond, Linda (Mar 2005). "Decline of an Island Fox Subspecies to Near Extinction". The Southwestern Naturalist. 50: 32–41. doi:10.1894/0038-4909(2005)050<0032:DOAIFS>2.0.CO;2.
- "Friends of the Island Fox: About Island Fox". www1.islandfox.org. Retrieved 2016-02-07.
- Kimura, Motoo (1962-06-01). "On the Probability of Fixation of Mutant Genes in a Population". Genetics. 47 (6): 713–719. ISSN 0016-6731. PMC 1210364. PMID 14456043.
- Haldane, J. B. S. (1927-07-01). "A Mathematical Theory of Natural and Artificial Selection, Part V: Selection and Mutation". Mathematical Proceedings of the Cambridge Philosophical Society. 23 (7): 838–844. Bibcode:1927PCPS...23..838H. doi:10.1017/S0305004100015644. ISSN 1469-8064.
- Fisher, R. A. (1990-01-01). "On the dominance ratio". Bulletin of Mathematical Biology. 52 (1–2): 297–318. doi:10.1007/BF02459576. ISSN 0092-8240. PMID 2185862.
- Hartwell, Leland (2011). Genetics: From Genes to Genomes. New York: McGraw-Hill. pp. 655–697. ISBN 978-0-07-352526-6.
- Kimura, Motoo (1983). The Neutral Theory of Molecular Evolution - Cambridge Books Online - Cambridge University Press. doi:10.1017/cbo9780511623486. ISBN 9780511623486.
- Molles, Manuel (2013). Ecology Concepts and Applications. New York: McGraw-Hill. ISBN 978-0-07353249-3.
- Broders, H. G.; Mahoney, S. P.; Montevecchi, W. A.; Davidson, W. S. (1999-08-01). "Population genetic structure and the effect of founder events on the genetic variability of moose, Alces alces, in Canada" (PDF). Molecular Ecology. 8 (8): 1309–1315. doi:10.1046/j.1365-294x.1999.00695.x. ISSN 0962-1083. PMID 10447871.
- Frankham, R. (1997-03-01). "Heredity - Abstract of article: Do island populations have less genetic variation than mainland populations?". Heredity. 78 (3): 311–327. doi:10.1038/hdy.1997.46. ISSN 0018-067X. PMID 9119706.
- Keller, Lukas F.; Waller, Donald M. (2002-05-01). "Inbreeding effects in wild populations". Trends in Ecology & Evolution. 17 (5): 230–241. doi:10.1016/S0169-5347(02)02489-8. ISSN 0169-5347.
- Özdil, Fulya; Ilhan, Fatma (21 July 2012). "DGAT1-exon8 polymorphism in Anatolian buffalo" (PDF). Livestock Science. 149 (1–2): 83–87. doi:10.1016/j.livsci.2012.06.030. Retrieved 6 Feb 2016.
- Habel, Jan Christian; Zachos, Frank Emmanuel; Finger, Aline; Meyer, Marc; Louy, Dirk; Assmann, Thorsten; Schmitt, Thomas (December 2009). "Unprecedented long-term genetic monomorphism in an endangered relict butterfly species". Conservation Genetics. 10 (6): 1659–1665. doi:10.1007/s10592-008-9744-5.