Flavobacteriia
The class Flavobacteriia is composed of a single class of environmental bacteria.[3] It contains the family Flavobacteriaceae, which is the largest family in the phylum Bacteroidota.[4] This class is widely distributed in soil, fresh, and seawater habitats.[5] The name is often spelt Flavobacteria, but was officially named Flavobacteriia in 2012.[6][7]
| Flavobacteriia | |
|---|---|
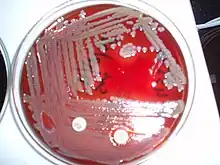 | |
| Elizabethkingia meningoseptica on blood agar | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Bacteroidota |
| Class: | Flavobacteriia Bernardet 2012[1] |
| Orders[2] | |
| |
Flavobacteriia are gram-negative aerobic rods, 2–5 μm long, 0.1–0.5 μm wide, with rounded or tapered ends.[6] They form circular cream to orange coloured colonies on agar, and are typically simple to successfully culture.[5] Flavobacteriia is a chemoorganotroph and are known for their ability to mineralize or degrade dissolved organic matter of high molecular weight and particulate plant material.[8]
Flavobacteriia have impacts on both the environment and human society, as they are able to cause diseases in many organisms. They are important in the decomposition of organic matter and pollutants, and are key members in the formation of marine biofilms.[9] They also have been known to cause diseases in some animal species, specifically bacterial cold water disease and columnaris disease.[10][11]
Taxonomy
Flavobacteriia is the largest of the four classes of phylum Bacteroidota. It is a single-order class, and its largest family is Flavobacteriaceae.[4] Flavobacteriaceae is the largest family in the phylum Bacteroidota. The family has over 90 genera and hundreds of species.[4] The genus Flavobacterium is most commonly used in studies of Flavobacteriia. This genus has 100 classified species with many additional unclassified species.[12] Recent taxonomic updates have reclassified several Flavobacterium species to new genera such as Microbacterium, Salegentibacter, and Planococcus.[13]
- Domain Bacteria
- Phylum Bacteroidota
- Class Flavobacteriia[12]
- Order Flavobacteriales[12]
- Family Crocinitomicaceae[12]
- Family Cryomorphaceae[12]
- Family Flavobacteriaceae[12]
- Genus Flavobacterium
- Genera Microbacterium, Salegentibacter, and Planococcus.[13]
- Family Ichthyobacteriaceae[12]
- Family Luteibaculaceae[12]
- Family Parvicellaceae[12]
- Family Salibacteraceae[12]
- Family Schleiferiaceae[12]
- Family Vicingaceae[12]
- Family Weeksellaceae[12]
- Family Flavobacteriales incertae sedis[12]
- Additionally, unclassified Flavobacteriales[12]
- Order Flavobacteriales[12]
- Class Flavobacteriia[12]
- Phylum Bacteroidota
History
.jpg.webp)
The genus Flavobacterium was established in 1889.[13] It was first written about in 1923 in Bergey's manual of determinative bacteriology and contained one of the first of 46 discussed species.[14] The manual defines Flavobacteriia as gram-negative, non-spore-forming, aerobic, non-gliding rods.[14] In 1999, Flavobacteriia was discovered to have a yellow pigment in colonies. It was also identified that they move through gliding and only grow in the presence of oxygen.[15]
In 1922, Flavobacterium columnare, an agent of columnaris disease with significant effects on fish, was discovered in the Mississippi River, making it one of the earliest known diseases of its kind.[11] The disease was originally labelled as a Myxobacteria in 1944, but was renamed to Flavobacterium columnare in 1996 with 10 species.[16][4] Flavobacteriia used to contain many non-related species of yellow, rod-shaped bacteria, but taxonomy has changed and stabilized due to the sequencing of rRNA to deduce phylogenetic relationships.[4]
Habitat

Flavobacteriia are widely distributed with high abundances in aquatic systems.[5] They have been found in diseased fish, microbial mats, freshwater and river sediments, seawater and marine sediments, soil, glaciers, and Antarctic lakes.[18][5] Increases in abundance are found in areas of high organic substrate inputs due to their role in the uptake, degradation, and decomposition of organic matter and can result in bacterial dominance.[13] Flavobacteriia is prominent in ocean sediments and decreases with increasing depth, and prefer sediments lacking vegetation.[19]
These bacteria also are highly abundant in melt ponds, solid ice cores, sea ice, and brine, as well as the photic zone.[18] More specifically, these photic zones show that Flavobacteriia are prominent in productive environments such as phytoplankton blooms and upwelling zones.[19] Flavobacteriia are prominent members of marine biofilms.[9] They have large impacts on the functioning of marine biofilms, however their abundance is believed to be heavily underestimated.[9]
Flavobacteriia can also be found in non-marine systems and are most common in Asian regions, specifically Korea and China, as well as Japan and India.[13]
Morphology
Flavobacteriia are a type of gram-negative rod-shaped bacteria with sizes typically ranging from 0.1μm to 0.5μm wide and 2μm to 5μm long.[6][20] Depending on the species of Flavobacteriia, the genome size can range from 1.85x109 daltons to 3.9x109 daltons.[21] Flavobacteriia are also unable to form endospores.[20] They are classified as gram-negative due to the composition of their cell wall, which consists of a thin layer of peptidoglycan surrounded by an outer membrane composed of lipopolysaccharides.[22] The rod-shape of these bacteria typically have straight or slightly curved parallel sides with rounded or slightly tapered ends.[6][8] The overall colony morphology of Flavobacteriia exhibit a circular shape that is either convex or slightly convex with a smooth appearance.[8] These colonies typically appear slightly translucent and can range in colour from pale yellow/cream to orange due to the presence of pigments such as carotenoids or flexirubin.[5][8]
Flavobacteriia do not possess flagella and rely on either a gliding motion or are non-motile.[21][23] The gliding motion allows them to move over wet surfaces such as a wet mount glass slide or agar plate.[23][24] Flavobacteriia exhibit a predominant forward gliding motion, but can also reverse direction and show flipping movements .[24] Research suggests that the gliding motion is facilitated by the proton gradient across the cytoplasmic membrane.[23][24]
Metabolism
Bacteria from the class Flavobacteriia have diverse metabolism. Flavobacteriia are chemoorganotrophic, meaning they use organic molecules as a source of energy.[8] Most species have obligately aerobic type of respiration, while some species can grow under weak microaerobic to anaerobic conditions.[8] Some species of Flavobacteriia have the ability to use a broad range of carbohydrates as energy sources, while others have a limited capacity or none at all, and instead prefer to utilize amino acids and proteins.[8] Approximately half of the species belonging to Flavobacteriia are capable of breaking down carbohydrates into acid and can degrade tyrosine and tween compounds.[8] Only a few species can degrade urea and DNA.[8] Many species also play a significant role in the mineralization of organic matter in both aquatic and soil environments due to their capability of breaking down various types of biomacromolecules.[8]
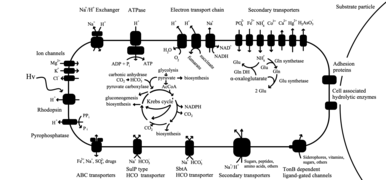
Flavobacteriia are not photosynthetic, but some marine species use proteorhodopsin for energy through the harvesting of light.[4] Proteorhodopsin (PR) is a proton pump that uses light, however species who use PR need to adapt to different environments to combat ultraviolet (UV) damage, and adopt the ability to mend DNA that has been damaged by UV.[25]
Proteorhodopsin is useful in the active transport of protons across the cell membrane. This is useful in the creation of ATP as energy in Flavobacteriia. The diagram to the right shows how proteorhodopsin is used in Flavobacteriia cells, and provides specifics about how it used in the bacteria.[25]
Culture
Typical culturing methods are used to isolate Flavobacteriia, such as simple dilutions. Techniques vary by species due to the high diversity of the class, however many are cultivated on simple media using yeast extract and a protein hydrolysate.[4] Sugars may need to be added or certain salts for marine species. Fish and bird pathogens may have additional requirements for culture methodology.[4]
Marine Flavobacteriia are cultured on marine agar or cytophaga agar. Non-marine Flavobacteriia are culture on rich media including nutrient agar, casitone-yeast extract agar, PYG agar, and TYES agar, or nutrient-poor media such as AO agar, PY2 agar, and R2A agar.[4] Flavobacteriia species that inhabit cold environments exhibit optimal growth at temperatures between 15 °C to 20 °C, while those that inhabit temperate environments exhibit optimal growth at temperatures between 20 °C to 30 °C.[8] Therefore, temperatures for culturing are between 20 °C and 30 °C, with an optimal temperature of 37˚C.[4]
Many psychrophilic and psychrotrophic species have been isolated through culture methods, mainly from polar regions.[4] Additional mesophilic species have been isolated as well as a few thermophiles, while extreme halophiles have not yet been identified.[4]
Environmental impact
Though the majority of Flavobacteriia are harmless, some infect opportunistically or cause severe diseases. This means that they can cause disease in many types organisms such as plants or fish.[13] They have proteins that discharge factors able to cause the development of a disease.[26] Fish pathogens are common on or in fish or the surrounding water. Bird pathogens cause outbreaks in domestic poultry or wild birds.[4]

One possible disease is bacterial cold water disease caused by Flavobacterium psychrophilum in rainbow trout, which can cause tissue erosion, jaw ulcerations, inflammation, and behavioural issues.[10] It can also cause acute losses in young rainbow trout, known as rainbow trout fry syndrome.[27] In 2005, the National Center for Cool and Cold Water Aquaculture measured survival rate to be 29.3% from these diseases.[27]
Additionally, Flavobacterium columare causes columnaris disease in freshwater fish species. Columnaris disease causes skin lesions, fin erosion, and gill necrosis, leading to mortality.[11]
Marine biofilms are a biological element that significantly affects the productivity and operation of marine habitats by assisting in basic microbial processes like photosynthesis, the cycling of nitrogen, and the degradation of organic matter and pollutants.[9] In the early stage of marine biofilms formation, Flavobacteriia colonize and form microcolonies to serve as a foundation for establishing other microorganisms in a community. In the biofilms community, Flavobacteriia also engage in a variety of cooperated interactions with other microbes rather than competition, including quorum sensing, nutrient sharing and scavenging. In sum, these interactions are essential for establishing and maintaining complex microbial communities in marine biofilms.[9]
Human impact
Food

Flavobacteriia have been linked to food and food product deterioration. The relative humidity of the shop where the product is located affects the growth of psychrophilic or psychrotrophic microorganisms.[13] Due to the formation of metabolic byproducts, spoilage of uncooked red flesh causes unpleasant smells, potential slime production, localised discolouration and unwanted flavours. Similarly, while Flavobacteriia are a continuous component of the initial flora in cold meats and fowl, they are unable to outcompete pseudomonads during preservation.[13] Poultry has a much greater prevalence of flavobacteriia than other fresh flesh.[13]
Flavobacteriia create pasteurisation-resistant extracellular enzymes, which causes the psychrotrophic deterioration of milk and dairy products.[13] Due to the creation of phospholipase C, they are also to blame for a decrease in cheddar cheese output and bitterness in milk. Given that phospholipases have the capacity to degrade the phospholipids that make up the milk fat globule membrane and thereby increase the vulnerability of the milk fat (triglycerides) to lipolytic assault, they may be significant in milk and milk products.[13]
Disease
Members of the Flavobacteriia also cause disease in humans.[13] However, as strains within the Flavobacterium were reclassified, many strains that cause human disease were transferred to new or different genera[28] such as Chryseobacterium, Myroides, Empedobacter and Sphingobacterium.[29] Their main infected populations are newborns and people with immunodeficiencies. Neonatal infections usually manifest as meningitis, and the mortality rate of neonatal meningitis is high. Meningitis can also cause bacteremia and pneumonia. In adults, infections can manifest in a variety of ways, including pneumonia, sepsis, meningitis, endocarditis, post-surgery, and post-burn.[29] To this point, existing pathogenic Flavobacteriia are currently very rare and difficult to detect, but remain a concern because they are resistant to many antimicrobial drugs.[28]
Industrial uses
The decomposition abilities of Flavobacteriia are also used to benefit humans industries. The bacteria commonly is found in sewage treatment facilities. They are used to treat wastewater because of their ability to digest chemicals and other molecules, including polycyclic aromatic hydrocarbons.[4]
Flavobacteriia is used to promote plant growth in the agricultural sector. The bacteria is able to solubilise inorganic phosphate and produce additional beneficial elements such as indole-3-acetic acid (the key plant hormone IAA) and 1-aminocyclopropane-1-carboxylatedeaminase (a hydrolase), which can be taken up and used by plants or alter their signalling.[30] It can act as a microbial agent to protect plants from other diseases, and even has benefits in the development of antimicrobial medicines.[4]
References
- Bernardet JF (2010). "Class II. Flavobacteriia class. nov.". In Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds.). Bergey's Manual of Systematic Bacteriology. Vol. 4 (2nd ed.). New York, NY: Springer. p. 105.
- Euzéby JP, Parte AC. "Flavobacteriia". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved June 29, 2021.
- Boone DR, Castenholz RW, eds. (2001). Bergey's Manual of Systematic Bacteriology. Vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria) (2nd ed.). New York: Springer-Verlag. pp. 465–466. ISBN 978-0-443-05615-4.
- McBride M (2014). "The Family Flavobacteriaceae". The Prokaryotes: 643–676. doi:10.1007/978-3-642-38954-2_130. ISBN 978-3-642-38953-5 – via SpringerLink.
- Buchan A, LeCleir GR, Gulvik CA, González JM (October 2014). "Master recyclers: features and functions of bacteria associated with phytoplankton blooms". Nature Reviews. Microbiology. 12 (10): 686–698. doi:10.1038/nrmicro3326. PMID 25134618. S2CID 26684717.
- Bernardet JF, Bowman JP (July 2013). "International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Flavobacterium and Cytophaga-like bacteria: Minutes of the meetings, 7 September 2011, Sapporo, Japan". International Journal of Systematic and Evolutionary Microbiology. 63 (Pt 7): 2752–2754. doi:10.1099/ijs.0.053926-0. PMID 23825377.
- "Flavobacteriia class". allmicrobes.com. Archived from the original on 1 November 2014. Retrieved 1 November 2014.
- Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, et al., eds. (2015-04-17). Bergey's Manual of Systematics of Archaea and Bacteria (1st ed.). Wiley. doi:10.1002/9781118960608.gbm00312. ISBN 978-1-118-96060-8. S2CID 242820836.
- Pollet T, Berdjeb L, Garnier C, Durrieu G, Le Poupon C, Misson B, Jean-François B (June 2018). "Prokaryotic community successions and interactions in marine biofilms: the key role of Flavobacteriia". FEMS Microbiology Ecology. 94 (6). doi:10.1093/femsec/fiy083. PMID 29733333.
- Barnes ME (2011). "A Review of Flavobacterium Psychrophilum Biology, Clinical Signs, and Bacterial Cold Water Disease Prevention and Treat" (PDF). The Open Fish Science Journal. 4 (1): 40–48. doi:10.2174/1874401X01104010040.
- Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, Decostere A (April 2013). "Columnaris disease in fish: a review with emphasis on bacterium-host interactions". Veterinary Research. 44 (1): 27. doi:10.1186/1297-9716-44-27. PMC 3648355. PMID 23617544.
- "Flavobacteriia". NCBI taxonomy database. National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine. Retrieved 2023-03-07.
- Waśkiewicz A, Irzykowska L (2014). "Flavobacterium spp. – Characteristics, Occurrence, and Toxicity". Encyclopedia of Food Microbiology: 938–942. doi:10.1016/B978-0-12-384730-0.00126-9. ISBN 978-0-12-384733-1.
- Loch TP, Faisal M (May 2015). "Emerging flavobacterial infections in fish: A review". Journal of Advanced Research. 6 (3): 283–300. doi:10.1016/j.jare.2014.10.009. PMC 4522593. PMID 26257926.
- "Species Flavobacterium aquatile". LPSN. Retrieved March 25, 2023.
- Kaiser D (2009). "Myxococcus". Encyclopedia of Microbiology (Third ed.): 220–244. doi:10.1016/B978-012373944-5.00018-3. ISBN 978-0-12-373944-5.
- Doghri I, Rodrigues S, Bazire A, Dufour A, Akbar D, Sopena V, Sablé S, Lanneluc I (October 2015). "Marine bacteria from the French Atlantic coast displaying high forming-biofilm abilities and different biofilm 3D architectures". BMC Microbiology. 15: 231. doi:10.1186/s12866-015-0568-4. PMC 4619314. PMID 26498445.
- Fernández-Gómez B, Díez B, Polz MF, Arroyo JI, Alfaro FD, Marchandon G, et al. (February 2019). "Bacterial community structure in a sympagic habitat expanding with global warming: brackish ice brine at 85-90 °N". The ISME Journal. 13 (2): 316–333. doi:10.1038/s41396-018-0268-9. PMC 6331608. PMID 30228379.
- Cleary DF, Coelho FJ, Oliveira V, Gomes NC, Polónia AR (2017). "Sediment depth and habitat as predictors of the diversity and composition of sediment bacterial communities in an inter-tidal estuarine environment". Marine Ecology. 38 (2): e12411. doi:10.1111/maec.12411.
- Zamora L, Vela AI, Sánchez-Porro C, Palacios MA, Domínguez L, Moore ER, Ventosa A, Fernández-Garayzábal JF (December 2013). "Characterization of flavobacteria possibly associated with fish and fish farm environment. Description of three novel Flavobacterium species: Flavobacterium collinsii sp. nov., Flavobacterium branchiarum sp. nov., and Flavobacterium branchiicola sp. nov". Aquaculture. 416–417: 346–353. doi:10.1016/j.aquaculture.2013.09.019.
- Shewan JM, McMeekin TA (October 1983). "Taxonomy (and ecology) of Flavobacterium and related genera". Annual Review of Microbiology. 37 (1): 233–252. doi:10.1146/annurev.mi.37.100183.001313. PMID 6357052.
- Lüderitz O, Freudenberg MA, Galanos C, Lehmann V, Rietschel ET, Shaw DH (January 1982). "Lipopolysaccharides of Gram-Negative Bacteria". Current Topics in Membranes and Transport. Vol. 17. Elsevier. pp. 79–151. doi:10.1016/s0070-2161(08)60309-3. ISBN 978-0-12-153317-5.
- Shrivastava A, Berg HC (December 2015). "Towards a model for Flavobacterium gliding". Current Opinion in Microbiology. 28: 93–97. doi:10.1016/j.mib.2015.07.018. PMC 4688146. PMID 26476806.
- McBride MJ, Zhu Y (January 2013). "Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes". Journal of Bacteriology. 195 (2): 270–278. doi:10.1128/JB.01962-12. PMC 3553832. PMID 23123910.
- Olson DK, Yoshizawa S, Boeuf D, Iwasaki W, DeLong EF (April 2018). "Proteorhodopsin variability and distribution in the North Pacific Subtropical Gyre". The ISME Journal. 12 (4): 1047–1060. doi:10.1038/s41396-018-0074-4. PMC 5864233. PMID 29476140.
- "Recent Advancements in Flavobacteria: Pathogenesis Mechanisms, Novel Genetic and Physiological Features, and Interaction with Hosts". Microbial Physiology and Metabolism – via Frontiers.
- Silverstein JT, Vallejo RL, Palti Y, Leeds TD, Rexroad CE, Welch TJ, et al. (March 2009). "Rainbow trout resistance to bacterial cold-water disease is moderately heritable and is not adversely correlated with growth" (PDF). Journal of Animal Science. 87 (3): 860–867. doi:10.2527/jas.2008-1157. PMID 19028851.
- García-López M, Santos J, Otero A (January 1999), "FLAVOBACTERIUM", in Robinson RK (ed.), Encyclopedia of Food Microbiology, Oxford: Elsevier, pp. 820–826, doi:10.1006/rwfm.1999.0660, ISBN 978-0-12-227070-3
- Booth J (January 2007), Enna SJ, Bylund DB (eds.), "Chryseobacterium and Related Genera Infections", xPharm: The Comprehensive Pharmacology Reference, New York: Elsevier, pp. 1–4, doi:10.1016/b978-008055232-3.62958-7, ISBN 978-0-08-055232-3
- Nishioka T, Elsharkawy MM, Suga H, Kageyama K, Hyakumachi M, Shimizu M (June 2016). "Development of Culture Medium for the Isolation of Flavobacterium and Chryseobacterium from Rhizosphere Soil". Microbes and Environments. 31 (2): 104–10. doi:10.1264/jsme2.ME15144. PMC 4912144. PMID 27098502.