Fluorescence cross-correlation spectroscopy
Fluorescence cross-correlation spectroscopy (FCCS) is a spectroscopic technique that examines the interactions of fluorescent particles of different colours as they randomly diffuse through a microscopic detection volume over time, under steady conditions.[1]
Discovery
Eigen and Rigler first introduced the fluorescence cross-correlation spectroscopy (FCCS) method in 1994. Later, in 1997, Schwille experimentally implemented this method.[2][3]
Theory
FCCS is an extension of the fluorescence correlation spectroscopy (FCS) method that uses two fluorescent molecules instead of one that emits different colours. The technique measures coincident green and red intensity fluctuations of distinct molecules that correlate if green and red labelled particles move together through a predefined confocal volume.[2] FCCS utilizes two species that are independently labeled with two different fluorescent probes of different colours. These fluorescent probes are excited and detected by two different laser light sources and detectors typically labeled as "green" and "red." By combining FCCS with a confocal microscope, the technique's capabilities are highlighted, as it becomes possible to detect fluorescence molecules in femtoliter volumes within the nanomolar range, with a high signal-to-noise ratio, and at a microsecond time scale.[4]
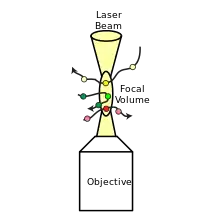
The normalized cross-correlation function is defined for two fluorescent species, G and R, which are independent green and red channels, respectively:
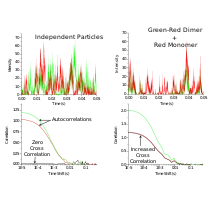
where differential fluorescent signals at a specific time, and at a delay time, later is correlated with each other. In the absence of spectral bleed-through -when the fluorescence signal from an adjacent channel is visible in the channel being observed-, the cross-correlation function is zero for non-interacting particles. In contrast to FCS, the cross-correlation function increases with increasing numbers of interacting particles.
FCCS is mainly used to study bio-molecular interactions both in living cells and in vitro.[5][2] It allows for measuring simple molecular stoichiometries and binding constants.[3] It is one of the few techniques that can provide information about protein-protein interactions at a specific time and location within a living cell. Unlike fluorescence resonance energy transfer, FCCS does not have a distance limit for interactions making it suitable for probing large complexes. However, FCCS requires active diffusion of the complexes through the microscope focus on a relatively short time scale, typically seconds.
Modeling
The mathematical function used to model cross-correlation curves in FCCS is slightly more complex compared to that used in FCS. One of the primary differences is the effective superimposed observation volume, denoted as in which the G and R channels form a single observation volume:
where and are radial parameters and and are the axial parameters for the G and R channels respectively.
The diffusion time, for a doubly (G and R) fluorescent species is therefore described as follows:
where is the diffusion coefficient of the doubly fluorescent particle.
The cross-correlation curve generated from diffusing doubly labelled fluorescent particles can be modelled in separate channels as follows:
In the ideal case, the cross-correlation function is proportional to the concentration of the doubly labeled fluorescent complex:
with
The cross-correlation amplitude is directly proportional to the concentration of double-labeled (red and green) species.[4][6]
Experimental method
FCCS measures the coincident green and red intensity fluctuations of distinct molecules that correlate if green and red labeled particles move together through a predefined confocal volume. To perform fluorescence cross-correlation spectroscopy (FCCS), samples of interest are first labeled with fluorescent probes of different colours. The FCCS setup typically includes a confocal microscope, two laser sources, and two detectors. The confocal microscope is used to focus the laser beams and collect the fluorescence signals. The signals from the detectors are then collected and recorded over time.[6][7] Data analysis involves cross-correlating the signals to determine the degree of correlation between the two fluorescent probes. This information can be used to extract data on the stoichiometry and binding constants of molecular complexes, as well as the timing and location of interactions within living cells.[6]
Applications
Fluorescence cross-correlation spectroscopy (FCCS) has several applications in the field of biophysics and biochemistry. Fluorescence cross-correlation spectroscopy (FCCS) is a powerful technique that enables the investigation of interactions between various types of biomolecules, including proteins, nucleic acids, and lipids.[8]
FCCS is one of the few techniques that can provide information about protein-protein interactions at a specific time and location within a living cell. FCCS can be used to study the dynamics of biomolecules in living cells, including their diffusion rates and localization.[9]
This can provide insights into the function and regulation of cellular processes. Unlike fluorescence resonance energy transfer, FCCS does not have a distance limit for interactions making it suitable for probing large complexes. However, FCCS requires active diffusion of the complexes through the microscope focus on a relatively short time scale, typically seconds. FCCS allows for measuring simple molecular stoichiometries and binding constants.[9]
References
- Thomas Weidemann; Petra Schwille (2013). "Fluorescence Cross-Correlation Spectroscopy". Encyclopedia of Biophysics. pp. 795–799. doi:10.1007/978-3-642-16712-6_509. ISBN 978-3-642-16711-9.
- Slaughter, B. D.; Unruh, J. R.; Li, R. Fluorescence fluctuation spectroscopy and imaging methods for examination of dynamic protein interactions in yeast. In Methods in Molecular Biology: Yeast Systems Biology. J.I. Castrillo and S.G. Oliver, Eds. (Springer, New York, 2011). Vol. 759, pp. 283-306.
- Chen, Y. and Mueller, J.D. Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy. (2006) Proc. Natl. Acad. Sci. U.S.A. 104, 3147-3152.
- Kirsten Bacia; Irina V. Majoul; Petra Schwille (August 2002). "Probing the Endocytic Pathway in Live Cells Using Dual-Color Fluorescence Cross-Correlation Analysis" (PDF). Biophysical Journal. 83 (2): 1184–1193. Bibcode:2002BpJ....83.1184B. doi:10.1016/S0006-3495(02)75242-9. PMC 1302220. PMID 12124298. Archived from the original (PDF) on 2015-09-24. Retrieved 2015-02-27.
- Bacia, K.; Kim, S.A.; Schwille, P. Fluorescence cross-correlation spectroscopy in living cells. (2006) Nat. Meth. 3, 83-89 .
- Laure Wawrezinieck; Hervé Rigneault; Didier Marguet; Pierre-François Lenne (December 2005). "Fluorescence Correlation Spectroscopy Diffusion Laws to Probe the Submicron Cell Membrane Organization". Biophysical Journal. 89 (6): 4029–4042. Bibcode:2005BpJ....89.4029W. doi:10.1529/biophysj.105.067959. PMC 1366968. PMID 16199500.
- Matthew J. Laurence; Timothy S. Carpenter; Ted A. Laurence; Matthew A. Coleman; Megan Shelby; Chao Liu (March 2022). "Biophysical Characterization of Membrane Proteins Embedded in Nanodiscs Using Fluorescence Correlation Spectroscopy". Membranes. 12 (4): 392. doi:10.3390/membranes12040392. PMC 9028781. PMID 35448362.
- Y Chen; J D Müller; K M Berland; E Gratton (October 1999). "Fluorescence fluctuation spectroscopy". Methods. 19 (2): 234–52. doi:10.1006/meth.1999.0854. PMID 10527729. S2CID 5609799.
- Kirsten Bacia; Elke Haustein; Petra Schwille (July 2014). "Fluorescence correlation spectroscopy: principles and applications". Cold Spring Harbor Protocols. 2014 (7): 709–25. doi:10.1101/pdb.top081802. PMID 24987147.
External links
- Fluorescence Cross Correlation (FCCS) (Becker & Hickl GmbH, web page)