Function-spacer-lipid Kode construct
Function-Spacer-Lipid (FSL) Kode constructs (Kode Technology) are amphiphatic, water dispersible biosurface engineering constructs that can be used to engineer the surface of cells, viruses and organisms, or to modify solutions and non-biological surfaces with bioactives.[1][2][3] FSL Kode constructs spontaneously and stably incorporate into cell membranes. FSL Kode constructs with all these aforementioned features are also known as Kode Constructs.[1] The process of modifying surfaces with FSL Kode constructs is known as "koding" and the resultant "koded" cells,[1] viruses and liposomes are respectively known as kodecytes,[1][4] and kodevirions.[1][5]
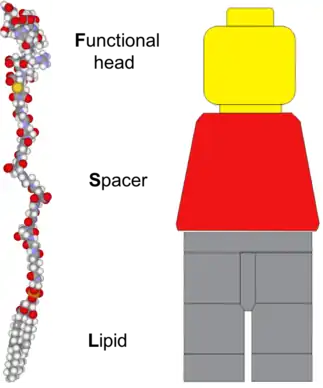
Technology description
All living surfaces are decorated with a diverse range of complex molecules, which are key modulators of chemical communications and other functions such as protection, adhesion, infectivity, apoptosis, etc. Functional-Spacer-Lipid (FSL) Kode constructs can be synthesized to mimic the bioactive components present on biological surfaces, and then re-present them in novel ways.[1][2]

The architecture of an FSL Kode construct, as implicit in the name, consists of three components - a functional head group, a spacer, and a lipid tail.[3] This structure is analogous to a Lego minifigure in that, they have three structural components, with each component having a separate purpose.[1][2] In the examples shown in all the figures, a Lego 'minifig' has been used for the analogy. However, it should be appreciated that this is merely a representation and the true structural similarity is significantly varied between Lego minifigures and FSL Kode constructs (fig 1). The functional group of an FSL is equivalent to a Lego minifigure head, with both being at the extremity and carrying the character functional components. The spacer of the FSL is equivalent to the body of the Lego minifigure and the arms on the minifigure are representative of substitutions which may be engineered into the chemical makeup of the spacer. The lipid of the FSL anchors it to lipid membranes and gives the FSL construct its amphiphatic nature which can cause it to self-assemble. Because the lipid tail can act directly as an anchor it is analogous to the legs of a Lego minifigure.
Flexible design
The functional group, the spacer and the lipid tail components of the FSL Kode construct can each be individually designed resulting in FSL Kode constructs with specific biological functions.[2] The functional head group is usually the bioactive component of the construct and the various spacers and lipids influence and effect its presentation, orientation and location on a surface. Critical to the definition of an FSL Kode construct is the requirement to be dispersible in water, and spontaneously and stably incorporate into cell membranes. Other lipid bioconjugates that include components similar to FSLs but do not have these features are not termed as Function-Spacer-Lipid Kode constructs.[2]
Functional groups[3]
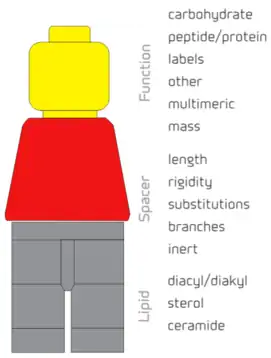
A large range of functional groups have already been made into FSL Kode constructs. These include:
- Carbohydrates – ranging from monosaccharides to polysaccharides and including blood group antigens,[1][2][6][7][8][9][10][11][12] hyaluronic acid oligomers and sialic acid residues[2]
- Peptide/protein – ranging from single amino acids[8][13][14][15] to proteins as large as antibodies[1]
- Labels – including fluorophores,[1][2][5][9][10][16][17] radioisotopes,[1][5] biotin,[1][2][9][10] etc.
- Other – chemical moieties such as maleimide,[15][18] click residues, PEG, charged compounds
Note 1: Multimeric – the presentation of the F residue can be as multimers with controlled spacing and be variable.
Note 2: Mass – the mass that can be anchored by an FSL Kode constructs can range from 200 to >1x106 Da
Spacers[3]
The spacer is an integral part of the FSL Kode construct and gives it several important characteristics including water dispersibility.[2]
- Length – the spacer can be varied in length, for example 1.9 nm (Ad), 7.2 nm (CMG2), 11.5 nm (CMG4),[2] allowing for enhanced presentation of Functional groups at the biosurface.
- Optimizes 'F' presentation – The presentation of the bioactive (functional group) on a spacer reduces steric hindrance and increases the bioactive surfaces exposed and available for interactions
- Rigidity – the spacer can be modified to be either flexible or rigid depending upon desired characteristics
- Substitutions (represented by the leaves on the stalk) – the spacer can be modified both in charge, and polarity.
- Branches – usually the spacer is linear, but it can also be branched including specific spacing of the branches to optimize presentation and interaction of the F group.
- Inert – important to the design of FSL Kode constructs is the biologically inert nature of the spacer. Importantly this feature means the S-L components of the constructs are unreactive with undiluted serum. Consequently, the constructs are compatible in vivo use, and can improve diagnostic assay sensitivity by allowing for the use of undiluted serum.
Lipids[3]
The lipid tail is essential for enabling lipid membrane insertion and retention but also for giving the construct amphiphilic characteristics that enable hydrophilic surface coating (due to formation of bilipid layers). Different membrane lipids that can be used to create FSLs have different membrane physiochemical characteristics and thus can affect biological function of the FSL. Lipids in FSL Kode constructs include:[2]
- Diacyl/diakyl e.g. DOPE
- Sterols e.g. cholesterol
- Ceramides
Optimising functional group (F) presentation

Upper image shows the antigen (flower head) presented in a variety of formats directly on a solid surface. The construct attaches randomly to the solid surface.
Lower image shows the same antigen (flower head) presented in optimal orientation as an FSL construct with a spacer (stalk) holding it away from the solid surface. The lipid tail (roots) causes the construct to attach to the solid surface.
In all diagrams, the same functional group is represented by the sunflower head. The first four examples show monomeric presentation of F, with the first example representing the short 1.9nm (Ad) spacer (the flower stalk). The second and fourth with the longer stalks (spacers) representing the CMG2 7.2nm spacer and CMG4 11.5nm spacers respectively. The second and third constructs have the same spacers, but the lipid tail of the third construct is cholesterol, rather than DOPE.
The last three examples all based on CMG2 spacers (stalks), with a DOPE lipid tail (roots) are multimeric presentations of F. The first of these is a simple duplication of the F group, while the following has an additional spacer between the F groups. The final construct is a cluster presentation.One of the important functions of an FSL construct is that it can optimise the presentation of antigens, both on cell surfaces and solid-phase membranes. This optimisation is achieved primarily by the spacer, and secondarily by the lipid tail. In a typical immunoassay, the antigen is deposited directly onto the microplate surface and binds to the surface either in a random fashion, or in a preferred orientation depending on the residues present on the surface of this antigen. Usually this deposition process is uncontrolled. In contrast, the FSL Kode construct bound to a microplate presents the antigen away from the surface in an orientation with a high level of exposure to the environment. Furthermore, typical immunoassays use recombinant peptides rather than discrete peptide antigens. As the recombinant peptide is many times bigger than the epitope of interest, a lot of undesired and unwanted peptide sequences are also represented on the microplate. These additional sequences may include unwanted microbial related sequences (as determined by a BLAST analysis) that can cause issues of low level cross-reactivity. Often the mechanism by which an immunoassay is able to overcome this low level activity is to dilute the serum so that the low level microbial reactive antibodies are not seen, and only high-level specific antibodies result in an interpretable result. In contrast, FSL Kode constructs usually use specifically selected peptide fragments (up to 40 amino acids),[15][18] thereby overcoming cross-reactivity with microbial sequences, and allowing for the use of undiluted serum (which increases sensitivity).
The F component can be further enhanced by presentation of it in multimeric formats and with specific spacing. The four types of multimeric format include linear repeating units, linear repeating units with spacing, clusters, and branching (Fig. 4).
Mechanisms of interaction



Amphiphilic FSL Kode construct
The FSL Kode construct by nature of its composition in possessing both hydrophobic and hydrophilic regions are amphiphilic (or amphipathic). This characteristic determines the way in which the construct will interact with surfaces. When present in a solution they may form simple micelles or adopt more complex bilayer structures with two simplistic examples shown in Fig. 5a. More complex structures are expected. The actual nature of FSL micelles has not been determined. However, based on normal structural function of micelles, it is expected that it will be determined in part by the combination of functional group, spacer and lipid together with temperature, concentration, size and hydrophobicity/hydrophilicity for each FSL Kode construct type.
Surface coatings will occur via two theoretical mechanisms, the first being direct hydrophobic interaction of the lipid tail with a hydrophobic surface resulting in a monolayer of FSL at the surface (Fig. 5b). Hydrophobic binding of the FSL will be via its hydrophobic lipid tail interacting directly with the hydrophobic (lipophilic) surface. The second surface coating will be through the formation of bilayers as the lipid tail is unable to react with the hydrophilic surface. In this case the lipids will induce the formation of a bilayer, the surface of which will be hydrophilic. This hydrophilic membrane will then interact directly with the hydrophilic surface and will probably encapsulate fibres. This hydrophilic bilayer binding is the expected mechanism by which FSLs are able to bind to fibrous membranes such as paper[19] and glass fibres (Fig. 5c) and (Fig. 9).
Lipid membrane modification
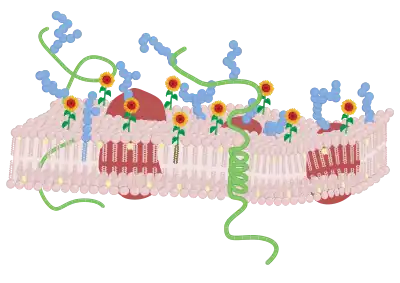

After labeling of the surface with the selected F bioactives, the constructs will be present and oriented at the membrane surface. It is expected that the FSL will be highly mobile within the membrane and the choice of lipid tail will effect is relative partitioning within the membrane.[2][7] The construct unless it has flip-flop behavior is expected to remain surface presented. However, the modification is not permanent in living cells and constructs will be lost (consumed) at a rate proportional to the activity at the membrane and division rate of the cell (with dead cells remaining highly labeled).[1] Additionally, when present in vivo with serum lipids FSLs will elute from the membrane into the plasma at a rate of about 1% per hour.[5][10] In fixed cells or inactive cells (e.g. red cells) stored in serum free media the constructs are retained normally.[1]
Liposomes are easy koded by simply adding FSL Kode constructs into the preparation. Contacting koded liposomes with microplates or other surfaces can cause the labeling of the microplate surface.
Non-biologic surface interaction
Non-biologic surface coatings will occur via two mechanisms, the first being direct hydrophobic interaction of the lipid tail with a hydrophobic surface resulting in a monolayer of FSL at the surface. The second surface coating will be through the formation of bilayers, which probably either encapsulate fibres or being via the hydrophilic F group. This is the expected mechanism by which FSLs bind to fibrous membranes such as paper[19] and glass fibres. A recent study has found that when FSL Kode constructs are optimised, could in a few seconds glycosylate almost any non-biological surface including metals, glass, plastics, rubbers, and other polymers.[20]
Technology features
The technological features of FSL Kode constructs and the koding process can be summarized as follows:
- Rapid and simple – simple contact for 10–120 minutes and constructs spontaneously and stably incorporate – no washing required.[1]
- Replicable – same variables (time, temperature, concentration) equals the same result.[1][6]
- Toxicity – FSL constructs are biocompatible, disperse into biological solutions without solvents, detergents. They label non-covalently and are non-genetic. Normal vitality and functionality is maintained in modified cells/virions/organisms.[7][9][10][16] Toxicity/vitality experiments in small laboratory animals, zebrafish, cell cultures, spermatozoa and embryos find no toxic effects within physiological ranges.
- Amphiphilic – the amphiphilic nature of the FSL Kode construct makes them water dispersible (clear solution of micelles), yet once interacted with a membrane they insert/coat and become water resistant
- Variable design – a single F can be presented in more than 100 ways by varying the spacer and lipid.
- High biovisibility – as the spacer holds the F moiety away for the membrane it is able to achieve increased sensitivity, specificity and reactivity can be optimized by use of multiple and variable biomarker presentations on the same surface.
- Additive – FSL modification is compatible with other technologies allowing users to add additional features to cells/viruses/organisms/surfaces already modified by more traditional methods. Multiple FSL constructs may be added to a surface simultaneously by simply creating a mix of FSL Kode constructs. Constructs insert into living or fixed cell (glutaraldehyde) membranes.
- Simple FSL peptide synthesis – there is a reactive-functional-group FSL Kode construct with maleimide as its functional group which can be used for preparation of FSLs from cysteine-containing peptides, proteins or any other thiols of biological interest.[15][18] The effective synthetic approach is based on the well-known Michael nucleophilic addition to maleimides (Fig. 7).
- Synthetic "Gylcolipids" – one family of the FSL constructs are synthetic glycolipids with well-defined hydrophobic tails and carbohydrate head groups
Koded membranes surfaces and solutions
FSL constructs have a wide range of uses and they have been used to modify the following:
- Cells – blood cells,[1][2][4][6][8][9][10][11][12][13][14][15][17][19][21] culture lines,[1] embryos,[1] spermatozoa[1]
- Viruses[5] – influenza,[2] measles, varicella
- Organisms – parasites, microbes, zebrafish[16]
- Liposomes – also micelles, lipid particles[9]
- Surfaces/fibres – hydrophobic or hydrophilic membranes/fibres, paper,[19] nitrocellulose, cotton, silk, glass, Teflon, silica, magnetic beads (microspheres)[22] etc.
- Solutions – saline, plasma/serum, culture media
Methodology for FSL use (koding)
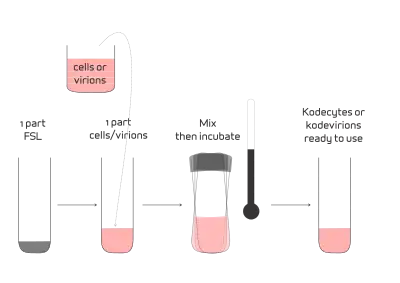
FSL constructs, when in solution (saline) and in contact, will spontaneously incorporate into cell and virus membranes.[1] The methodology involves simply preparing a solution of FSL constructs in the range of 1–1000 μg/mL. The actual concentration will depend on the construct and the quantity of construct required in the membrane. One part of FSL solution is added to one part of cells (up to 100% suspension) and they are incubated at a set temperature within the range of 4–37 °C (39–99 °F) depending on temperature compatibility of the cells being modified. The higher the temperature, the faster the rate of FSL insertion into the membrane. For red blood cells, at 37 °C incubation for 2 hours achieves >95% insertion with at least 50% insertion being achieved within 20 minutes. In general, FSL insertion time of 4 hours at room temperature or 20 hours at 4 °C gives results similar to 1 hour at 37 °C for carbohydrate based FSLs inserting into red blood cells.[1] The resultant kodecytes or kodevirions do not required to be washed, however this option should be considered if an excess of FSL construct is used in the koding process.
Applications
FSL Kode constructs have been used for research and development, diagnostic products, and are currently being investigated as potential therapeutic agents.
Kodecytes
FSL have been used to create human red cell kodecytes that have been used to detect and identify blood group allo-antibodies[13][14][15] as ABO sub-group mimics,[11] ABO quality control systems,[4] serologic teaching kits[12] and a syphilis diagnostic.[21] Kodecytes have also demonstrated that FSL-FLRO4 is a suitable reagent for labelling packed red blood cells (PRBC) at any point during routine storage and look to facilitate the development of immunoassays and transfusion models focused on addressing the mechanisms involved in tansfusion-related immunomodulation (TRIM).[17] Murine kodecytes have been experimentally used to determine in vivo cell survival,[10] and create model transfusion reactions.[9][10] Zebrafish kodecytes have been used to determine real time in vivo cell migration.[16] Kodecytes have been used to create influenza diagnostics.[2] Kodecytes which have been modified with FSL-GB3 were unable to be infected with the HIV virus.[7][23]
Kodevirions
Kodevirions are FSL modified viruses. Several FSL Kode constructs have been used to label viruses to assist in their flow-cytometric visualisation[5] and to track them real time distribution in animal models.[5] They have also been used to modify the surface of viruses with the intention of targeting them to be used to attach tumors (oncolytic).[5]
Kodesomes
Kodesomes are liposomes that have been decorated with FSL Kode constructs. These have been used to deposit FSL constructs onto microplates to create diagnostic assays. They also have the potential for therapeutic use.[24]
Koded solutions
These are solutions containing FSL Kode constructs where the construct will exist as a clear micellular dispersion. FSL-GB3 as a solution/gel has been used to inhibit HIV infection[7] and to neutralise Shiga toxin.[7] FSL blood group A as a solution has been used to neutralise circulating antibodies in a mouse model and allow incompatible blood group A (murine kodecytes) transfusion.[9] This model experiment was used to demonstrate the potential of FSLs to neutralise circulating antibody and allow for incompatible blood transfusion or organ transplantation.[19]
Koded surfaces

All FSL Kode constructs disperse in water and are therefore compatible with inkjet printers. FSL constructs can be printed with a standard desktop inkjet printer directly onto paper to create immunoassays.[19] An empty ink cartridge is filled with an FSL construct and words, barcodes, or graphics are printed. A Perspex template is adhered to the surface to create reaction wells. The method is then a standard EIA procedure, but blocking of serum is not required and undiluted serum can be used. A typical procedure is as follows: add serum, incubate, wash by immersion, add secondary EIA conjugate, incubate, wash, add NBT/BCIP precipitating substrate and stop the reaction when developed by washing (Fig. 9). The end result is stable for years.
See also
External links
- FSL Constructs: A Simple Method for Modifying Cell/Virion Surfaces with a Range of Biological Markers Without Affecting their Viability – Journal of Visualised Experiments (JOVE) free video article
- kodecyte.org - the academic resource for Kode Technology
References
- Blake, Debbie A; Bovin, Nicolai V; Bess, Dan; Henry, Stephen M (2011). "FSL Constructs: A Simple Method for Modifying Cell/Virion Surfaces with a Range of Biological Markers Without Affecting their Viability". Journal of Visualized Experiments. 54 (e3289). doi:10.3791/3289. PMC 3211133. PMID 21847082.
- Korchagina, Elena; Tuzikov, Alexander; Formanovsky, Andrey; Popova, Inna; Henry, Stephen; Bovin, Nicolai (2012). "Toward creating cell membrane glycolandscapes with glycan lipid constructs". Carbohydrate Research. 356: 238–46. doi:10.1016/j.carres.2012.03.044. PMID 22551471.
- Korchagina, E. Y.; Henry, S. M. (2015-07-16). "Synthetic glycolipid-like constructs as tools for glycobiology research, diagnostics, and as potential therapeutics". Biochemistry (Moscow). 80 (7): 857–871. doi:10.1134/S0006297915070068. ISSN 0006-2979. PMID 26542000. S2CID 14965044.
- Henry, Stephen M (2009). "Modification of red blood cells for laboratory quality control use". Current Opinion in Hematology. 16 (6): 467–472. doi:10.1097/MOH.0b013e328331257e. PMID 19680123. S2CID 37416831.
- Hadac, Elizabeth M; Federspiel, Mark J; Cherneyy, Evgeny; Tuzikov, Alexander; Korchagina, Elena; Bovin, Nicolai V; Russel, Stephen; Henry, Stephen M (2011). "Fluorescein and radiolabeled Function-Spacer-Lipid constructs allow for simple in vitro and in vivo bioimaging of enveloped virions". Journal of Virological Methods. 176 (1–2): 78–84. doi:10.1016/j.jviromet.2011.06.005. PMID 21703308.
- Frame, Tom; Carroll, Tim; Korchagina, Elena; Bovin, Nicolai; Henry, Stephen (2007). "Synthetic glycolipid modification of red blood cell membranes". Transfusion. 47 (5): 876–882. CiteSeerX 10.1.1.494.2776. doi:10.1111/j.1537-2995.2007.01204.x. PMID 17465953. S2CID 18086433.
- Harrison, Amanda L; Olsson, Martin L; Brad Jones, R; Ramkumar, Stephanie; Sakac, Darinka; Binnington, Beth; Henry, Stephen; Lingwood, Clifford A; Branch, Donald R (2010). "A synthetic globotriaosylceramide analogue inhibits HIV-1 infection in vitro by two mechanisms". Glycobiology. 27 (5): 515–524. doi:10.1007/s10719-010-9297-y. PMID 20582467. S2CID 1422452.
- Georgakopoulos, T; Komarraju, Sarvani; Henry, Stephen; Bertolini, Joseph (2011). "An improved Fc function assay utilising CMV antigen coated red blood cells generated with synthetic Function-Spacer-Lipid constructs". Vox Sanguinis. 102 (1): 72–78. doi:10.1111/j.1423-0410.2011.01512.x. PMID 21749406. S2CID 9758322.
- Oliver, Caroline; Blake, Debbie; Henry, Stephen (2011). "In vivo neutralization of anti-A and successful transfusion of A antigen incompatible red cells in an animal model". Transfusion. 51 (12): 2664–2675. doi:10.1111/j.1537-2995.2011.03184.x. PMID 21599675. S2CID 205724219.
- Oliver, Caroline; Blake, Debbie; Henry, Stephen (2011). "Modeling transfusion reactions and predicting in vivo cell survival with kodecytes". Transfusion. 51 (8): 1723–1730. doi:10.1111/j.1537-2995.2010.03034.x. PMID 21303367. S2CID 24736518.
- Hult, Annika K; Frame, Tim; Chesla, Scott; Henry, Stephen; Olsson, Martin L (2012). "Flow cytometry evaluation of red blood cells mimicking naturally-occurring ABO subgroups following modification with variable amounts of FSL-A and B constructs". Transfusion. 52 (2): 247–251. doi:10.1111/j.1537-2995.2011.03268.x. PMID 21812783. S2CID 5984970.
- Henry, Stephen; Perry, Holly (2012). FSL-A+B(tri) Serologic Teaching Kit Technical Bulletin. Scholarly Commons. hdl:10292/2827.
- Heathcote, Damien; Carrol, Tim; Wang, Jui-Jen; Flower, Robert; Rodionov, Igor; Tuzikov, Alexander; Bovin, Nicolai; Henry, Stephen (2010). "Novel antibody screening cells, MUT+Mur kodecytes, created by attaching peptides onto erythrocytes". Transfusion. 50 (3): 635–641. doi:10.1111/j.1537-2995.2009.02480.x. PMID 19912581. S2CID 20952307.
- Nadarajan, V.S.; Laing, A. A.; Saad, S. M.; Usin, M (2011). "Prevalence and specificity of red-blood-cell antibodies in a multiethnic South and East Asian patient population and influence of using novel MUT+Mur+ kodecytes on its detection". Vox Sanguinis. 102 (1): 65–71. doi:10.1111/j.1423-0410.2011.01507.x. PMID 21592136. S2CID 20297050.
- Henry, Stephen; Komarraju, Sarvani; Heathcote, Damien; Rodinov, Igor L (2011). "Designing peptide-based FSL constructs to create Miltenberger kodecytes". ISBT Science Series. 6 (2): 306–312. doi:10.1111/j.1751-2824.2011.01505.x.
- Lan, C-C; Blake, D; Henry, S; Love, D R (2012). "Fluorescent Function-Spacer-Lipid construct labelling allows for real-time in vivo imaging of cell migration and behaviour in zebrafish (Danio rerio)". Journal of Fluorescence. 22 (4): 1055–63. doi:10.1007/s10895-012-1043-3. PMID 22434405. S2CID 14406691.
- Ki, Katrina K.; Flower, Robert L.; Faddy, Helen M.; Dean, Melinda M. (Jan 7, 2016). "Incorporation of fluorescein conjugated function-spacer-lipid constructs into the red blood cell membrane facilitates detection of labeled cells for the duration of ex-vivo storage" (PDF). Journal of Immunological Methods. 429: 66–70. doi:10.1016/j.jim.2016.01.003. PMID 26773455.
-
Henry, Stephen; Rodionov, Igor (2012). "FSL-RFG(Maleimide) FSL Construction Kit Technical Bulletin". Scholarly Commons. hdl:10292/2241.
{{cite journal}}: Cite journal requires|journal=(help) - Henry, Stephen; Barr, Katie; Oliver, Caroline (2012). "Modeling transfusion reactions with kodecytes and enabling ABO incompatible transfusion with Function-Spacer-Lipid constructs". ISBT Science Series. 7 (1): 106–111. doi:10.1111/j.1751-2824.2012.01563.x. S2CID 73310504.
- Williams, Eleanor; Barr, Katie; Korchagina, Elena; Tuzikov, Alexander; Henry, Stephen; Bovin, Nicolai (2016-01-16). "Ultra-Fast Glyco-Coating of Non-Biological Surfaces". International Journal of Molecular Sciences. 17 (1): 118. doi:10.3390/ijms17010118. PMC 4730359. PMID 26784187.
- Chesla, S; Henry, S; Eatz, R; Sinor, L (2010). "Solid phase syphilis test utilizing KODE technology". Transfusion. 50: 196A–197A. doi:10.1111/j.1537-2995.2010.02833_1.x. PMID 20815863.
- "Rapid biofunctionalization of magnetic beads with function-spacer-lipid constructs". sepmag.eu. Retrieved 2015-12-01.
- Branch, D R; Harrison, A; Sakac, D; Lingwood, C; Henry, S (2008). "A synthetic, water dispersible glycolipid analogue of the Pk blood group antigen completely blocks HIV-1 infection". Transfusion. 48 (2S): 1–325. doi:10.1111/j.1537-2995.2008.01891.x. hdl:10292/4136. PMID 18783347. S2CID 222199165.
- Harrison, A L; Henry, S; Mahfoud, R; Manis, A; Albertini, A; Gaudin, Y; Lingwood, C A; Branch, D R (2010). "A novel VSV/HIV pseudotype approach for the study of HIV microbicides without requirement for level 3 biocontainment". Future Virology. 6 (10): 1241–1259. doi:10.2217/fvl.11.88.