GCM transcription factors
In molecular biology, the GCM transcription factors are a family of proteins which contain a GCM motif. The GCM motif is a domain that has been identified in proteins belonging to a family of transcriptional regulators involved in fundamental developmental processes which comprise Drosophila melanogaster GCM and its mammalian homologues (human GCM1 and GCM2).[1][2][3][4] In GCM transcription factors the N-terminal moiety contains a DNA-binding domain of 150 amino acids. Sequence conservation is highest in this GCM domain. In contrast, the C-terminal moiety contains one or two transactivating regions and is only poorly conserved.
| GCM | |||||||||
|---|---|---|---|---|---|---|---|---|---|
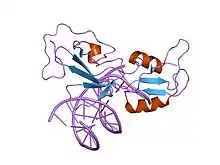 structure of the gcm domain bound to dna | |||||||||
| Identifiers | |||||||||
| Symbol | GCM | ||||||||
| Pfam | PF03615 | ||||||||
| Pfam clan | CL0274 | ||||||||
| InterPro | IPR003902 | ||||||||
| PROSITE | PS50807 | ||||||||
| SCOP2 | 1odh / SCOPe / SUPFAM | ||||||||
| |||||||||
The GCM motif has been shown to be a DNA binding domain that recognises preferentially the nonpalindromic octamer 5'-ATGCGGGT-3'.[1][2][3] The GCM motif contains many conserved basic amino acid residues, seven cysteine residues, and four histidine residues.[1] The conserved cysteines are involved in shaping the overall conformation of the domain, in the process of DNA binding and in the redox regulation of DNA binding.[3] The GCM domain as a new class of Zn-containing DNA-binding domain with no similarity to any other DNA-binding domain.[5] The GCM domain consists of a large and a small domain tethered together by one of the two Zn ions present in the structure. The large and the small domains comprise five- and three-stranded beta-sheets, respectively, with three small helical segments packed against the same side of the two beta-sheets. The GCM domain exercises a novel mode of sequence-specific DNA recognition, where the five-stranded beta-pleated sheet inserts into the major groove of the DNA. Residues protruding from the edge strand of the beta-pleated sheet and the following loop and strand contact the bases and backbone of both DNA strands, providing specificity for its DNA target site.
References
- Akiyama Y, Hosoya T, Poole AM, Hotta Y (December 1996). "The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals". Proc. Natl. Acad. Sci. U.S.A. 93 (25): 14912–6. Bibcode:1996PNAS...9314912A. doi:10.1073/pnas.93.25.14912. PMC 26236. PMID 8962155.
- Schreiber J, Sock E, Wegner M (April 1997). "The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain". Proc. Natl. Acad. Sci. U.S.A. 94 (9): 4739–44. Bibcode:1997PNAS...94.4739S. doi:10.1073/pnas.94.9.4739. PMC 20794. PMID 9114061.
- Schreiber J, Enderich J, Wegner M (May 1998). "Structural requirements for DNA binding of GCM proteins". Nucleic Acids Res. 26 (10): 2337–43. doi:10.1093/nar/26.10.2337. PMC 147556. PMID 9580683.
- Tuerk EE, Schreiber J, Wegner M (February 2000). "Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMb". J. Biol. Chem. 275 (7): 4774–82. doi:10.1074/jbc.275.7.4774. PMID 10671510.
- Cohen SX, Moulin M, Hashemolhosseini S, Kilian K, Wegner M, Muller CW (April 2003). "Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition". EMBO J. 22 (8): 1835–45. doi:10.1093/emboj/cdg182. PMC 154474. PMID 12682016.