Gene-environment interplay
Gene-environment interplay is a term encompassing multiple ways that genes and environments work together to produce phenotypic traits. Processes classified as examples of gene-environment interplay include gene-environment interaction, gene-environment correlation,[1] and epigenetics.[2] It is often studied with behavioral genetic research designs like twin, family, and adoption studies.[1]
Aim
The aim of studying gene-environment interplay is to discover new mechanisms of disease and to describe the reasons for digression from the expected expression of disease.[3] Studying these interactions allows for the complete understanding of diseases that are inclusive of numerous discrete and interacting pathways.[3] By better understanding the etiology of such diseases, interventions can be designed to target individual factors within specific populations that minimize the prevalence of disease.
Types
Gene-environment interplay may be complementary or antagonistic. Complementary interactions refer to both environmental exposure and genetic predisposition affecting disease risk in the same manner (i.e. both increasing or decreasing).[3] An example is atherosclerosis, which is a multifactorial disease, influenced by both genetic and environmental factors. The allele for the familial hypercholesterolemia (FH) gene increases the risk of atherosclerosis by enhancing the build-up of low-density lipoprotein (LDL) cholesterol. Large intakes of saturated fats also lend themselves to increasing low-density lipoprotein cholesterol levels. Both complementary elements act to augment atherosclerosis susceptibility. Antagonistic interactions are when the two factors are opposing (i.e. one increases risk, while the other decreases it).[3] An example is hypertension. Hypertension susceptibility is increased through the presence of known risk alleles, while consistent physical activity acts as an antagonist to decrease the likelihood of developing hypertension.[3]
Environmental Factors
Pollutants
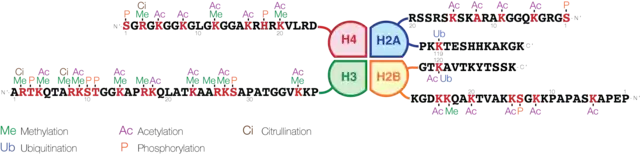
Epigenetic modifications, such as DNA methylation (DNAm), acetylation, histone tail modifications (such as methylation and acetylation), phosphorylation, ubiquitylation, microRNA (miRNA) expression can affect gene activity.[4][5] Decreased DNA methylation is thought to be linked to air pollution exposure. The mechanism behind this is not completely understood, but it may involve the formation of reactive oxygen species. These species generate oxidative stress, which stimulates downstream signalling.[5] This can lead to hydroxymethylation, which replaces the methyl group with a hydroxyl group. Hydroxymethylation alters gene expression, potentially causing diseases such as lung cancer.[5] Air pollution can also decrease expression of DNA methyltransferases, which would decrease overall DNA methylation.[5] This can ultimately lead to inflammation in regions such as the airways, which triggers asthma. Pathways can also be activated to increase cytokine expression and immune cells in the same regions.[5]
Malnutrition
Nutrition can affect gene expression, thereby altering phenotype. Fetal starvation has been linked to decreased DNA methylation levels, particularly on the imprinted IGF2 loci associated with insulin metabolism.[6] This can increase the risk for metabolic disorders and type II diabetes mellitus.[7] Studies on prenatal exposure to famine have discovered that malnutrition causes differential DNA methylation of genes associated with growth, development and metabolism, increasing the risk of adverse phenotypes such as obesity and high cholesterol later in life.[8]
Exercise
Physical activity increases telomerase activity[9] and induces epigenetic modifications of specific genes. For example, it has been shown to increase methylation of the ASC gene, which generally decreases with age, reducing the expression of pro-inflammatory cytokines.[10] This suppression can help prevent the development of chronic inflammation and associated age-related diseases due to excess inflammatory cytokines.[10] However, these epigenetic modifications depend on the intensity and type of exercise and are reversible with the cessation of physical activity.[11]
Studying Gene-Environment Interplay
Adoption and Twin Studies
Adoption and twin studies have been critical in revealing the interplay between genotype and environment. These studies have contributed to studies of behaviour, personality, and psychiatric illnesses.[12] A Finnish adoption study on schizophrenia revealed that a healthy environment can mitigate the effects of genetics in adopted individuals born to schizophrenic mothers.[13] Criminal and antisocial behaviour have also been found to be influenced by both genetic and environmental factors through these types of studies.[12][14]
Medical Conditions
Gene-environment interplay has been found to play a part in the majority of diseases. For instance, gene-environment interactions have a prevalent role in mental health disorders; specifically, evidence has found a link to alcohol dependence,[15] schizophrenia,[16] and psychosis.[17] There is a common polymorphism in the AKT1 gene that causes its carriers to be more sensitive to developing psychosis with consistent cannabis use.[17] Evidence also supports gene-environment interplay to be connected to cardiovascular and metabolic conditions.[3] These include roles in obesity,[18] pulmonary disease,[19] and diabetes.[20] The rise in the incidence of diabetes is suggested to be linked to interactions between the FTO or KCNQ1 genes and environmental factors.[20]
References
- Neiderhiser, Jenae M.; Liu, Chang; Griffin, Amanda M. (2018). "Gene–Environment Interplay". In Bornstein, Marc H. (ed.). The SAGE Encyclopedia of Lifespan Human Development. 2455 Teller Road, Thousand Oaks, California 91320: SAGE Publications, Inc. doi:10.4135/9781506307633.n349. ISBN 9781506307657.
{{cite encyclopedia}}: CS1 maint: location (link) - Sokolowski, Maria B.; Goodwin, Stephen F. (2012-09-13). Gene-Environment Interplay. Academic Press. pp. ix. ISBN 9780123876874.
- Flowers, E., Froelicher, E.S., & Aouizerat, B.E. (2012). Gene-environment interactions in cardiovascular disease. European Journal of Cardiovascular Nursing, 11(4), 472–478. https://doi.org/10.1016/j.ejcnurse.2011.06.001
- Weinhold, B. (2006). Epigenetics: The science of change. Environmental Health Perspectives, 114(3), A160-A167. https://doi.org/10.1289/ehp.114-a160
- Rider, C. F., & Carlsten, C. (2019). Air pollution and DNA methylation: Effects of exposure in humans. Clinical Epigenetics, 11(1), 131-131. https://doi.org/10.1186/s13148-019-0713-2
- Heijmans, B. T., Tobi, E. W., Stein, A. D., Putter, H., Blauw, G. J., Susser, E. S., Slagboom, P. E., & Lumey, L. H. (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences, 105(44), 17046–17049. https://doi.org/10.1073/pnas.0806560105
- Tiffon, C. (2018). The impact of nutrition and environmental epigenetics on human health and disease. International Journal of Molecular Sciences, 19(11), 3425. https://doi.org/10.3390/ijms19113425
- Tobi, E. W., Slieker, R. C., Stein, A. D., Suchiman, H. E. D., Slagboom, P. E., van Zwet, E. W., Heijmans, B. T., & Lumey, L. (2015). Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. International Journal of Epidemiology, 44(4), 1211–1223. https://doi.org/10.1093/ije/dyv043
- Werner, C. M., Hecksteden, A., Morsch, A., Zundler, J., Wegmann, M., Kratzsch, J., Thiery, J., Hohl, M., Bittenbring, J. T., Neumann, F., Böhm, M., Meyer, T., & Laufs, U. (2018). Differential effects of endurance, interval, and resistance training on telomerase activity and telomere length in a randomized, controlled study. European Heart Journal, 40(1), 34–46. https://doi.org/10.1093/eurheartj/ehy585
- Nakajima, K., Takeoka, M., Mori, M., Hashimoto, S., Sakurai, A., Nose, H., Higuchi, K., Itano, N., Shiohara, M., Oh, T., & Taniguchi, S. (2010). Exercise Effects on Methylation of ASC Gene. International Journal of Sports Medicine, 31(09), 671–675. https://doi.org/10.1055/s-0029-1246140
- Sellami, M., Bragazzi, N., Prince, M. S., Denham, J., & Elrayess, M. (2021). Regular, Intense Exercise Training as a Healthy Aging Lifestyle Strategy: Preventing DNA Damage, Telomere Shortening and Adverse DNA Methylation Changes Over a Lifetime. Frontiers in Genetics, 12. https://doi.org/10.3389/fgene.2021.652497
- Atkinson, B.E. & Vernon, P.A. (2020). Gene–Environment Interaction. In The Wiley Encyclopedia of Personality and Individual Differences (eds B.J. Carducci, C.S. Nave, A. Fabio, D.H. Saklofske and C. Stough). 207-210. https://doi.org/10.1002/9781118970843.ch212
- Tienari, P., Wynne, L. C., Moring, J., Lahti, I., Naarala, M., Sorri, A., Wahlberg, K. E., Saarento, O., Seitamaa, M., Kaleva, M., & Läksy, K. (1994). The Finnish Adoptive Family Study of Schizophrenia. British Journal of Psychiatry, 164(S23), 20–26. https://doi.org/10.1192/s0007125000292696
- Cadoret, R. J., Cain, C. A., & Crowe, R. R. (1983b). Evidence for gene-environment interaction in the development of adolescent antisocial behavior. Behavior Genetics, 13(3), 301–310. https://doi.org/10.1007/bf01071875
- van der Zwaluw, C.S., & Engels, R.C. (2009). Gene-environment interactions and alcohol use and dependence: current status and future challenges. Addiction, 104(6), 907-914. https://doi.org/10.1111/j.1360-0443.2009.02563.x
- van Os, J., Rutten, B.P., & Poulton, R. (2008). Gene-environment interactions in schizophrenia: review of epidemiological findings and future directions. Schizophrenia Bulletin, 34(6), 1066-1082. https://doi.org/10.1093/schbul/sbn117
- Zwicker, A., Denovan-Wright, E.M., & Uher, R. (2018). Gene-environment interplay in the etiology of psychosis. Psychological Medicine, 48(12), 1925-1936. https://doi.org/10.1017/s003329171700383x
- Andreasen, C.H., & Andersen, G. (2009). Gene-environment interactions and obesity--further aspects of genomewide association studies. Nutrition, 25(10), 998-1003. https://doi.org/10.1016/j.nut.2009.06.001
- Hirvonen, A. (2009). Gene-environment interactions in chronic pulmonary diseases. Mutation Research, 667(1-2), 132-141. https://doi.org/10.1016/j.mrfmmm.2008.12.013
- Kido, Y. (2017). Gene-environment interaction in type 2 diabetes. Diabetology International, 8(1), 7-13. https://doi.org/10.1007/s13340-016-0299-2