Colletotrichum lindemuthianum
Colletotrichum lindemuthianum is a fungus which causes anthracnose, or black spot disease, of the common bean plant (Phaseolus vulgaris). It is considered a hemibiotrophic pathogen because it spends part of its infection cycle as a biotroph, living off of the host but not harming it, and the other part as a necrotroph, killing and obtaining nutrients from the host tissues.
| Colletotrichum lindemuthianum | |
|---|---|
 | |
| Colletotrichum lindemuthianum damage to bean pods. | |
| Scientific classification | |
| Domain: | Eukaryota |
| Kingdom: | Fungi |
| Division: | Ascomycota |
| Class: | Sordariomycetes |
| Order: | Glomerellales |
| Family: | Glomerellaceae |
| Genus: | Colletotrichum |
| Species: | C. lindemuthianum |
| Binomial name | |
| Colletotrichum lindemuthianum (Sacc. & Magnus) Briosi & Cavara, (1889) | |
| Synonyms | |
|
Gloeosporium lindemuthianum Sacc. & Magnus, (1878) | |
History
The anthracnose of common bean was first identified in 1875 in the fruit and vegetable garden of the Agricultural Institute of Popplesdorf, Germany by Lindemuth.[1] By 1878, Saccardo and Magnus had made many observations on the cause of the anthracnose disease, recording their results in Michelia I:129.[2] They concluded that it was caused by a fungus, which they named Gloeosporium lindemuthianum after Lindemuth himself.[1] Several years later, Briosi and Cavara discovered the presence of setae on the fungus, reclassifying it from the genus Gloeosporium to Colletotrichum, where it remains today.[1][2] Recognizing the devastating effect the fungus was having on common bean populations worldwide, it quickly became a heavily studied subject among scientists, who principally investigated means of controlling its spread. In 1911, Barrus reported the discovery of multiple fungal strains, each of which differed in its ability to infect certain varieties of bean plants, which initiated the work of Edgerton and Moreland, who found eleven different strains of the pathogen, but theorized more may exist.[1] Since then, numerous strains have been identified, each targeting specific varieties of bean plants. During the early part of the 20th century, the various races were identified by use of the Greek alphabet, paired with numbers, but at the turn of the 21st century a naming system using binary code was adopted.[3] Under the binary naming system, each plant cultivar is given a binary number, and the code for a particular race of the pathogen is determined by the sum of the binary numbers of the cultivars which it infects.[3]
Life cycle
Infection
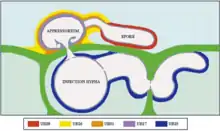
The spores of C. lindemuthianum are dispersed by rain splash and must quickly attach to the aerial parts of the plant in order to infect the host.[4] A heavy rainfall may spread the spores as far as 4.5 meters from the host plant.[3] The spore will then germinate on the new host and form a short germ tube which develops an appressorium, or 'pressing' organ.[5] As the germ tube grows, it pulls the spore and the appressorium together, causing an indentation to occur in the cell wall. An infection peg is then able to protrude from the appressorium and penetrate through the cell wall. Once through the cell wall, an infection hypha grows and develops into an infection vesicle.[1][6]
Biotrophic phase
The first stage after infection is known as the biotrophic phase, and consists of a broad primary hyphae, which develops out of the infection vesicle. The primary hyphae occasionally penetrates through additional cell walls by use of mechanical force, but usually will not grow very far from the infection vesicle. It always stays along a wall, such that half of the hyphae's circumference is in contact with the cell wall at all times. The primary hyphae do not penetrate the host cell's plasma membranes, but instead grows between it and the cell wall. As such, these hyphae do not intentionally kill any cells.[1][7] In these early stages of infection, proteins are released by the infection vesicle which suppress the host's defense responses. One such protein is formed by the nitrogen starvation-induced gene CgDN3. The proteins suppress any hypersensitivity responses from the host to allow unhindered growth and development of the fungus. During the biotrophic phase, the pathogen gains nutrients by transferring hexoses and amino acids from the living host cell to the fungus by use of monosaccharide-H+ symporters.[7]
Necrotrophic phase
About 48–72 hours after inoculation, depending on the environment, the biotrophic phase ends and the necrotrophic phase begins. It is marked by the development of many thin hyphae, known as secondary or necrotrophic hyphae, which develop off of the primary hyphae and the infection hypha. Unlike the much larger primary hyphae, these secondary hyphae move freely through the host in all directions, penetrating cell walls and membranes alike.[7] In order to more easily ramify throughout the host tissue, the hyphae release enzymes which break down host-specific proteins.[6] Some such enzymes include cutinase, proteases, pectin and pectate lyases. One enzyme, endopolygalacturonase, is a highly specialized cell wall-degrading enzyme which is critical for the growth of the mycelium. Endopolygalacturonase is normally produced by fruiting plants and induces ripening of fruit by degrading polygalacturonan present in the cell walls. C. lindemuthianum releases large amounts of this enzyme, which not only weakens the cell wall by removing the polygalacturonan, ng the area available for cell-wall degradation by other enzymes.[8] All of this cell degradation is not allowed to go to waste, however, as the breaking down of the cell wall releases many oligo and monosaccharides which are then free to be taken up by the fungus. This is a much broader availability of sugars than was available in the biotrophic phase, which consisted mostly of glucose and fructose derived from sucrose. Because of how rapidly the fungus is killing the host, it no longer releases any defense response prevention proteins, relying purely on the rapid death of the host and growth of the pathogen.[7] Additionally, since the mycelium must rapidly spread during the necrotrophic stage in order to supply nutrients to the fungus, it will more easily thrive in younger bean plants, which have softer tissues than their older counterparts.[1] While there is very little discoloration of the host cells before secondary hyphae development, discoloration becomes rapidly apparent about 100 hours post infection.[1] Black spots begin to develop on the surface of the plant, and grows radially outward: the manifestation of anthracnose disease. These are the first visual symptoms of an infected plant, and usually occur along leaf veins on the underside of the leaf. As the lesions grow, they become indented in the center, where conidia begin to develop. These conidia are colorless at first, but develop into light pink, flesh-colored pustules ready to be spread to new hosts by rain. In the event of infection of a bean pod, the conidia may develop in the seeds themselves, where they can remain dormant until the seed begins to germinate and grow, at which point the fungus will begin to grow on the young host.[9]
Conidial anastomosis tubes
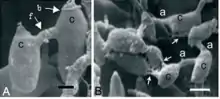
Maturing colonies Colletotrichum lindemuthianum, produce conidial anastomosis tubes (CATs) between conidia before leaving the host and before germination,[10] in contrast to the genetic model fungus Neurospora crassa (bread mould) that produces conidial anastomosis tubes from conidia and conidial germ tubes during germination.[11] Other Colletotrichum species produce conidial anastomosis tubes as well.[10] The initial characterization of CATs was made in this plant pathogen in 2003 by Roca et al. in Brazil.[10][12] CATs first form 15 days after the start of fruiting body development and grow from the conidia. As they grow, they may either fuse with other CATs, as evidenced by fusion points, or make direct contact with another conidium. Because each conidium creates CATs, the result is a network of conidia all connected together. The CAT network is developed very quickly, often within 1 hour of the first CAT connection. The exact purpose of this connection network is unclear, but it permits free flow of cytoplasm, proteins, organelles and even nuclei between conidia. It is unclear whether these CATs can play a role in genetic recombination, as no sexual stages for C. lindemuthianum has been found in nature.[10]
Economic effects

Dry bean is an ancient crop, first domesticated over 7000 years ago and is still a staple crop globally.[3] Today, the largest bean producing nations are Brazil, India, China, Mexico, The United States of America, Myanmar, Canada, and Argentina. In all of these nations, anthracnose is considered one of the most invasive and destructive dry bean diseases, capable of destroying up to 95% of a plantation's yield while also threatening growth and development rates. As such, it is very important for producers to have disease-free certified seeds, which are generally produced in arid regions such as Idaho, where the relative humidity does not surpass the necessary 92% for spore germination. Further precaution is often taken by producers, who apply fungicides such as Azoxystrobin, fluodiooxonil or metalaxyl-m. Applied at a concentration of 125 grams per hectare of dry bean crop, azoxystrobin has been shown to increase bean yields by up to 20%.[3]
Growing in culture
It has been known for some time that the fungus can be grown in a culture. The culture is generally made of a nutrient rich agar, as the fungus generally has a hard time germinating in water due to the water solubility of the appressorium. For optimal growth, the culture should be kept at 22 °C and at a pH of 8, although it can be grown in temperatures between 0–34 °C and pH levels between 3–11.[1]
Diseases on fungus
In 1975, British mycologist Rawlinson published findings of many isometric, uniformly sized particles that he identified as double-stranded RNA viruses in the extract of the α5 race of fungus. The α5 race is noted for its unusually poor sporulation in cultures and its weak pathogenicity. Rawlinson hypothesized these features were caused by the observed viral particles.[13] However, the particles proved to be inseparable from the fungus, making an effective control difficult to acquire. Instead, he compared growth, pathogenicity and morphology of the α5 race with other races of C. lindemuthianum which were not infected. No significant differences were observed between the infected and non-infected races of the pathogen, although all α races proved to be highly nonpathogenic, regularly scoring under 1 on the 5 point pathogenicity scale, whereas other races, such as the δ races, had pathogenicity ratings of 5. While the virus has no observable negative effects on the fungus, its purpose or how it infected the fungus is still unknown.[13]
References
- Leach, Julian Gilbert. 1922.The parasitism of colletotrichum lindemuthianum. University of Minnesota. Retrieved 30 March 2014
- Stoneman, Bertha. 1898. A comparative study of the development of some anthracnoses. Botanical Gazette 26 (2) (August, 1898).
- Pynenburg, Gerard Martin. 2010. Argonomic and economic assessment of intensive pest management of dry bean. M.Sc., University of Guelph. Retrieved 4 April 2014
- Mercure, E.W.; H. Kunoh; R.L. Nicholson (December 1994). "Adhesion of Colletotrichum graminicola conidia to corn leaves, a requirement for disease development". Physiological and Molecular Plant Pathology. 45 (6): 407. doi:10.1016/S0885-5765(05)80039-6.
- Bailey, J.A. (1992). Colletotrichum: biology, pathology and control. p. 88. Archived from the original on 2014-04-13. Retrieved 2013-05-08.
- Dean, Ralph; VanKan, Jan A. L.; Pretorius, Zacharias A.; Hammond-Kosack, Kim E.; Di Pietro, Antonio; Spanu, Pietro De.; Rudd, Jason J.; Dickman, Marty; Kahmann, Regine; Ellis, Jeff; Foster, Gary D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology. p. 414. Accessed 31 March 2014.
- Munch, Steffen, Ulrike Lingner, Daniela S. Floss, Nancy Ludwig, Norbert Sauer, and Holger B. Deising. 2008. The hemibiotrophic lifestyle of colletotrichum species. Elsevier(165): 41.
- Acosta-rodríguez, Ismael, Carlos Piñón-escobedo, Ma Guadalupe Zavala-páramo, Everardo López-romero, and Horacio Cano-camacho. 2005. Degradation of cellulose by the bean-pathogenic fungus colletotrichum lindemuthianum. production of extracellular cellulolytic enzymes by cellulose induction. Antonie van Leeuwenhoek 87, (4) (05): 301-10
- Saettler, A. February 01, 1983. Bean anthracnose Seed-transmitted disease caused by the fungus Colletotrichum lindemuthianum.. Extension bulletin E - Cooperative Extension Service, Michigan State University. no. 1671, (accessed March 27, 2014).
- Roca M., M.G.; Davide, L.C.; Mendes-Costa, M.C.; Wheals, A. (2003). "Conidial anastomoses tubes in Colletotrichum". Fungal Genetics and Biology. 40 (2): 138–145. doi:10.1016/S1087-1845(03)00088-4. PMID 14516766.
- Roca, M.G.; Arlt, J.; Jeffree, C.E.; Read, N.D. (2005). "Cell biology of conidial anastomosis tubes in Neurospora crassa". Eukaryotic Cell. 4 (5): 911–919. doi:10.1128/EC.4.5.911-919.2005. PMC 1140100. PMID 15879525.
- Glass, N.L.; Fleissner, A. (2006). "Re-wiring the network: understanding the mechanism of function of anastomosis in filamentous fungi.". In Kues, U.; Fisher, R. (eds.). Growth, Differentiation and Sexuality (The Mycota. Mycota. Vol. 1. Springer. pp. 123–139. ISBN 978-3-540-28134-4.
- RAWLINSON, CJ. 1975. DOUBLE-STRANDED-RNA VIRUS IN COLLETOTRICHUM-LINDEMUTHIANUM. Transactions of the British Mycological Society 65, no. oct, (accessed April 01, 2014).
Further reading
Roca M., M. Gabriela; Davide, Lisete C.; Mendes-Costa, Maria C. Cytogenetics of Colletotrichum lindemuthianum (Glomerella cingulata f. sp. phaseoli) Fitopatologia brasileira, vol. 28 no. 4 Brasília July/Aug. 2003