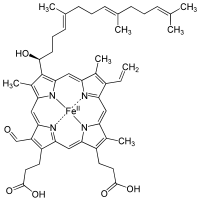
Heme
Heme (American English), or haem (Commonwealth English, both pronounced /hi:m/ HEEM), is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.[1]

In biochemical terms, heme is a coordination complex "consisting of an iron ion coordinated to a porphyrin acting as a tetradentate ligand, and to one or two axial ligands."[2] The definition is loose, and many depictions omit the axial ligands.[3] Among the metalloporphyrins deployed by metalloproteins as prosthetic groups, heme is one of the most widely used[4] and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigment in blood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and endothelial nitric oxide synthase.[5][6]
The word haem is derived from Greek αἷμα haima meaning "blood".

Function
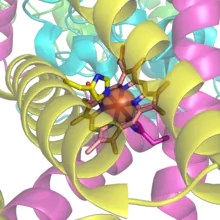
Hemoproteins have diverse biological functions including the transportation of diatomic gases, chemical catalysis, diatomic gas detection, and electron transfer. The heme iron serves as a source or sink of electrons during electron transfer or redox chemistry. In peroxidase reactions, the porphyrin molecule also serves as an electron source, being able to delocalize radical electrons in the conjugated ring. In the transportation or detection of diatomic gases, the gas binds to the heme iron. During the detection of diatomic gases, the binding of the gas ligand to the heme iron induces conformational changes in the surrounding protein.[7] In general, diatomic gases only bind to the reduced heme, as ferrous Fe(II) while most peroxidases cycle between Fe(III) and Fe(IV) and hemeproteins involved in mitochondrial redox, oxidation-reduction, cycle between Fe(II) and Fe(III).
It has been speculated that the original evolutionary function of hemoproteins was electron transfer in primitive sulfur-based photosynthesis pathways in ancestral cyanobacteria-like organisms before the appearance of molecular oxygen.[8]
Hemoproteins achieve their remarkable functional diversity by modifying the environment of the heme macrocycle within the protein matrix.[9] For example, the ability of hemoglobin to effectively deliver oxygen to tissues is due to specific amino acid residues located near the heme molecule.[10] Hemoglobin reversibly binds to oxygen in the lungs when the pH is high, and the carbon dioxide concentration is low. When the situation is reversed (low pH and high carbon dioxide concentrations), hemoglobin will release oxygen into the tissues. This phenomenon, which states that hemoglobin's oxygen binding affinity is inversely proportional to both acidity and concentration of carbon dioxide, is known as the Bohr effect.[11] The molecular mechanism behind this effect is the steric organization of the globin chain; a histidine residue, located adjacent to the heme group, becomes positively charged under acidic conditions (which are caused by dissolved CO2 in working muscles, etc.), releasing oxygen from the heme group.[12]
Types
Major hemes
There are several biologically important kinds of heme:
| Heme A | Heme B | Heme C | Heme O | ||
|---|---|---|---|---|---|
| PubChem number | 7888115 | 444098 | 444125 | 6323367 | |
| Chemical formula | C49H56O6N4Fe | C34H32O4N4Fe | C34H36O4N4S2Fe | C49H58O5N4Fe | |
| Functional group at C3 |  |
–CH(OH)CH2Far | –CH=CH2 | –CH(cystein-S-yl)CH3 | –CH(OH)CH2Far |
| Functional group at C8 | –CH=CH2 | –CH=CH2 | –CH(cystein-S-yl)CH3 | –CH=CH2 | |
| Functional group at C18 | –CH=O | –CH3 | –CH3 | –CH3 | |
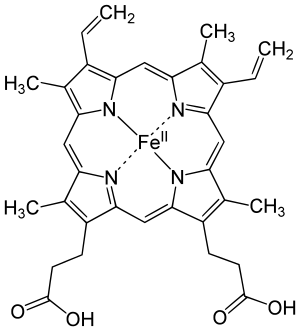
The most common type is heme B; other important types include heme A and heme C. Isolated hemes are commonly designated by capital letters while hemes bound to proteins are designated by lower case letters. Cytochrome a refers to the heme A in specific combination with membrane protein forming a portion of cytochrome c oxidase.[15]
Other hemes
- The following carbon numbering system of porphyrins is an older numbering used by biochemists and not the 1–24 numbering system recommended by IUPAC which is shown in the table above.
- Heme l is the derivative of heme B which is covalently attached to the protein of lactoperoxidase, eosinophil peroxidase, and thyroid peroxidase. The addition of peroxide with the glutamyl-375 and aspartyl-225 of lactoperoxidase forms ester bonds between these amino acid residues and the heme 1- and 5-methyl groups, respectively.[16] Similar ester bonds with these two methyl groups are thought to form in eosinophil and thyroid peroxidases. Heme l is one important characteristic of animal peroxidases; plant peroxidases incorporate heme B. Lactoperoxidase and eosinophil peroxidase are protective enzymes responsible for the destruction of invading bacteria and virus. Thyroid peroxidase is the enzyme catalyzing the biosynthesis of the important thyroid hormones. Because lactoperoxidase destroys invading organisms in the lungs and excrement, it is thought to be an important protective enzyme.[17]
- Heme m is the derivative of heme B covalently bound at the active site of myeloperoxidase. Heme m contains the two ester bonds at the heme 1- and 5-methyl groups also present in heme l of other mammalian peroxidases, such as lactoperoxidase and eosinophil peroxidase. In addition, a unique sulfonamide ion linkage between the sulfur of a methionyl amino-acid residue and the heme 2-vinyl group is formed, giving this enzyme the unique capability of easily oxidizing chloride and bromide ions to hypochlorite and hypobromite. Myeloperoxidase is present in mammalian neutrophils and is responsible for the destruction of invading bacteria and viral agents. It perhaps synthesizes hypobromite by "mistake". Both hypochlorite and hypobromite are very reactive species responsible for the production of halogenated nucleosides, which are mutagenic compounds.[18][19]
- Heme D is another derivative of heme B, but in which the propionic acid side chain at the carbon of position 6, which is also hydroxylated, forms a γ-spirolactone. Ring III is also hydroxylated at position 5, in a conformation trans to the new lactone group.[20] Heme D is the site for oxygen reduction to water of many types of bacteria at low oxygen tension.[21]
- Heme S is related to heme B by having a formal group at position 2 in place of the 2-vinyl group. Heme S is found in the hemoglobin of a few species of marine worms. The correct structures of heme B and heme S were first elucidated by German chemist Hans Fischer.[22]
The names of cytochromes typically (but not always) reflect the kinds of hemes they contain: cytochrome a contains heme A, cytochrome c contains heme C, etc. This convention may have been first introduced with the publication of the structure of heme A.
Use of capital letters to designate the type of heme
The practice of designating hemes with upper case letters was formalized in a footnote in a paper by Puustinen and Wikstrom[23] which explains under which conditions a capital letter should be used: "we prefer the use of capital letters to describe the heme structure as isolated. Lowercase letters may then be freely used for cytochromes and enzymes, as well as to describe individual protein-bound heme groups (for example, cytochrome bc, and aa3 complexes, cytochrome b5, heme c1 of the bc1 complex, heme a3 of the aa3 complex, etc)." In other words, the chemical compound would be designated with a capital letter, but specific instances in structures with lowercase. Thus cytochrome oxidase, which has two A hemes (heme a and heme a3) in its structure, contains two moles of heme A per mole protein. Cytochrome bc1, with hemes bH, bL, and c1, contains heme B and heme C in a 2:1 ratio. The practice seems to have originated in a paper by Caughey and York in which the product of a new isolation procedure for the heme of cytochrome aa3 was designated heme A to differentiate it from previous preparations: "Our product is not identical in all respects with the heme a obtained in solution by other workers by the reduction of the hemin a as isolated previously (2). For this reason, we shall designate our product heme A until the apparent differences can be rationalized.".[24] In a later paper,[25] Caughey's group uses capital letters for isolated heme B and C as well as A.
Synthesis

The enzymatic process that produces heme is properly called porphyrin synthesis, as all the intermediates are tetrapyrroles that are chemically classified as porphyrins. The process is highly conserved across biology. In humans, this pathway serves almost exclusively to form heme. In bacteria, it also produces more complex substances such as cofactor F430 and cobalamin (vitamin B12).[26]
The pathway is initiated by the synthesis of δ-aminolevulinic acid (dALA or δALA) from the amino acid glycine and succinyl-CoA from the citric acid cycle (Krebs cycle). The rate-limiting enzyme responsible for this reaction, ALA synthase, is negatively regulated by glucose and heme concentration. Mechanism of inhibition of ALAs by heme or hemin is by decreasing stability of mRNA synthesis and by decreasing the intake of mRNA in the mitochondria. This mechanism is of therapeutic importance: infusion of heme arginate or hematin and glucose can abort attacks of acute intermittent porphyria in patients with an inborn error of metabolism of this process, by reducing transcription of ALA synthase.[27]
The organs mainly involved in heme synthesis are the liver (in which the rate of synthesis is highly variable, depending on the systemic heme pool) and the bone marrow (in which rate of synthesis of Heme is relatively constant and depends on the production of globin chain), although every cell requires heme to function properly. However, due to its toxic properties, proteins such as Hemopexin (Hx) are required to help maintain physiological stores of iron in order for them to be used in synthesis.[28] Heme is seen as an intermediate molecule in catabolism of hemoglobin in the process of bilirubin metabolism. Defects in various enzymes in synthesis of heme can lead to group of disorder called porphyrias, these include acute intermittent porphyria, congenital erythropoetic porphyria, porphyria cutanea tarda, hereditary coproporphyria, variegate porphyria, erythropoietic protoporphyria.[29]
Synthesis for food
Impossible Foods, producers of plant-based meat substitutes, use an accelerated heme synthesis process involving soybean root leghemoglobin and yeast, adding the resulting heme to items such as meatless (vegan) Impossible burger patties. The DNA for leghemoglobin production was extracted from the soybean root nodules and expressed in yeast cells to overproduce heme for use in the meatless burgers.[30] This process claims to create a meaty flavor in the resulting products.[31][32]
Degradation

Degradation begins inside macrophages of the spleen, which remove old and damaged erythrocytes from the circulation.
In the first step, heme is converted to biliverdin by the enzyme heme oxygenase (HO).[33] NADPH is used as the reducing agent, molecular oxygen enters the reaction, carbon monoxide (CO) is produced and the iron is released from the molecule as the ferrous ion (Fe2+).[34] CO acts as a cellular messenger and functions in vasodilation.[35]
In addition, heme degradation appears to be an evolutionarily-conserved response to oxidative stress. Briefly, when cells are exposed to free radicals, there is a rapid induction of the expression of the stress-responsive heme oxygenase-1 (HMOX1) isoenzyme that catabolizes heme (see below).[36] The reason why cells must increase exponentially their capability to degrade heme in response to oxidative stress remains unclear but this appears to be part of a cytoprotective response that avoids the deleterious effects of free heme. When large amounts of free heme accumulates, the heme detoxification/degradation systems get overwhelmed, enabling heme to exert its damaging effects.[28]
| heme | heme oxygenase-1 | biliverdin + Fe2+ | |
 |
 | ||
| H+ + NADPH + O2 | NADP+ + CO | ||
 | |||
In the second reaction, biliverdin is converted to bilirubin by biliverdin reductase (BVR):[37]
| biliverdin | biliverdin reductase | bilirubin | |
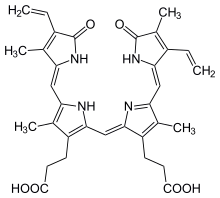 |
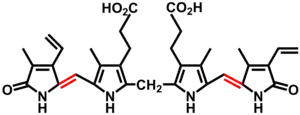 | ||
| H+ + NADPH | NADP+ | ||
 | |||
Bilirubin is transported into the liver by facilitated diffusion bound to a protein (serum albumin), where it is conjugated with glucuronic acid to become more water-soluble. The reaction is catalyzed by the enzyme UDP-glucuronosyltransferase.[38]
| bilirubin | UDP-glucuronosyltransferase | bilirubin diglucuronide | |
 |
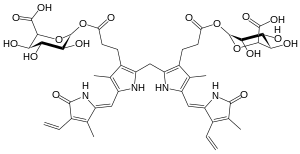 | ||
| 2 UDP-glucuronide | 2 UMP + 2 Pi | ||
 | |||
This form of bilirubin is excreted from the liver in bile. Excretion of bilirubin from liver to biliary canaliculi is an active, energy-dependent and rate-limiting process. The intestinal bacteria deconjugate bilirubin diglucuronide and convert bilirubin to urobilinogens. Some urobilinogen is absorbed by intestinal cells and transported into the kidneys and excreted with urine (urobilin, which is the product of oxidation of urobilinogen, and is responsible for the yellow colour of urine). The remainder travels down the digestive tract and is converted to stercobilinogen. This is oxidized to stercobilin, which is excreted and is responsible for the brown color of feces.[39]
In health and disease
Under homeostasis, the reactivity of heme is controlled by its insertion into the “heme pockets” of hemoproteins. Under oxidative stress however, some hemoproteins, e.g. hemoglobin, can release their heme prosthetic groups.[40][41] The non-protein-bound (free) heme produced in this manner becomes highly cytotoxic, most probably due to the iron atom contained within its protoporphyrin IX ring, which can act as a Fenton's reagent to catalyze in an unfettered manner the production of free radicals.[42] It catalyzes the oxidation and aggregation of protein, the formation of cytotoxic lipid peroxide via lipid peroxidation and damages DNA through oxidative stress. Due to its lipophilic properties, it impairs lipid bilayers in organelles such as mitochondria and nuclei.[43] These properties of free heme can sensitize a variety of cell types to undergo programmed cell death in response to pro-inflammatory agonists, a deleterious effect that plays an important role in the pathogenesis of certain inflammatory diseases such as malaria[44] and sepsis.[45]
Cancer
There is an association between high intake of heme iron sourced from meat and increased risk of colon cancer.[46] The heme content of red meat is 10 times higher than that of white meat such as chicken.[47]
The American Institute for Cancer Research (AICR) and World Cancer Research Fund International (WCRF) concluded in a 2018 report that there is limited but suggestive evidence that foods containing heme iron increase risk of colorectal cancer.[48] A 2019 review found that heme iron intake is associated with increased breast cancer risk.[49]
Genes
The following genes are part of the chemical pathway for making heme:
- ALAD: aminolevulinic acid, δ-, dehydratase (deficiency causes ala-dehydratase deficiency porphyria)[50]
- ALAS1: aminolevulinate, δ-, synthase 1
- ALAS2: aminolevulinate, δ-, synthase 2 (deficiency causes sideroblastic/hypochromic anemia)
- CPOX: coproporphyrinogen oxidase (deficiency causes hereditary coproporphyria)[51]
- FECH: ferrochelatase (deficiency causes erythropoietic protoporphyria)
- HMBS: hydroxymethylbilane synthase (deficiency causes acute intermittent porphyria)[52]
- PPOX: protoporphyrinogen oxidase (deficiency causes variegate porphyria)[53]
- UROD: uroporphyrinogen decarboxylase (deficiency causes porphyria cutanea tarda)[54]
- UROS: uroporphyrinogen III synthase (deficiency causes congenital erythropoietic porphyria)
Notes and references
- Bloomer, Joseph R. (1998). "Liver metabolism of porphyrins and haem". Journal of Gastroenterology and Hepatology. 13 (3): 324–329. doi:10.1111/j.1440-1746.1998.01548.x. PMID 9570250. S2CID 25224821.
- Chemistry, International Union of Pure and Applied (2009). "Hemes (heme derivatives)". IUPAC Compendium of Chemical Terminology. doi:10.1351/goldbook.H02773. ISBN 978-0-9678550-9-7. Archived from the original on 22 August 2017. Retrieved 28 April 2018.
{{cite book}}:|website=ignored (help) - A standard biochemistry text defines heme as the "iron-porphyrin prosthetic group of heme proteins"(Nelson, D. L.; Cox, M. M. "Lehninger, Principles of Biochemistry" 3rd Ed. Worth Publishing: New York, 2000. ISBN 1-57259-153-6.)
- Poulos, Thomas L. (2014-04-09). "Heme Enzyme Structure and Function". Chemical Reviews. 114 (7): 3919–3962. doi:10.1021/cr400415k. ISSN 0009-2665. PMC 3981943. PMID 24400737.
- Paoli, M. (2002). "Structure-function relationships in heme-proteins" (PDF). DNA Cell Biol. 21 (4): 271–280. doi:10.1089/104454902753759690. hdl:20.500.11820/67200894-eb9f-47a2-9542-02877d41fdd7. PMID 12042067. S2CID 12806393. Archived (PDF) from the original on 2018-07-24.
- Alderton, W.K. (2001). "Nitric oxide synthases: structure, function and inhibition". Biochem. J. 357 (3): 593–615. doi:10.1042/bj3570593. PMC 1221991. PMID 11463332.
- Milani, M. (2005). "Structural bases for heme binding and diatomic ligand recognition in truncated hemoglobins". J. Inorg. Biochem. 99 (1): 97–109. doi:10.1016/j.jinorgbio.2004.10.035. PMID 15598494.
- Hardison, R. (1999). "The Evolution of Hemoglobin: Studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story". American Scientist. 87 (2): 126. doi:10.1511/1999.20.809.
- Poulos, T. (2014). "Heme Enzyme Structure and Function". Chem. Rev. 114 (7): 3919–3962. doi:10.1021/cr400415k. PMC 3981943. PMID 24400737.
- Thom, C. S. (2013). "Hemoglobin Variants: Biochemical Properties and Clinical Correlates". Cold Spring Harbor Perspectives in Medicine. 3 (3): a011858. doi:10.1101/cshperspect.a011858. PMC 3579210. PMID 23388674.
- Bohr; Hasselbalch, Krogh. "Concerning a Biologically Important Relationship - The Influence of the Carbon Dioxide Content of Blood on its Oxygen Binding". Archived from the original on 2017-04-18.
{{cite journal}}: Cite journal requires|journal=(help) - Ackers, G. K.; Holt, J. M. (2006). "Asymmetric cooperativity in a symmetric tetramer: human hemoglobin". J. Biol. Chem. 281 (17): 11441–3. doi:10.1074/jbc.r500019200. PMID 16423822. S2CID 6696041.
- Caughey, W. S.; Smythe, G. E.; O'Keeffe, D. H.; Maskasky, J. E.; Smith, M. L. (1975). "Heme A of Cytochrome c Oxidase: Structure and properties: comparisons with hemes B, C, and S and derivatives". J. Biol. Chem. 250 (19): 7602–7622. doi:10.1016/S0021-9258(19)40860-0. PMID 170266.
- Hegg, Eric L. (2004). "Heme A Synthase Does Not Incorporate Molecular Oxygen into the Formyl Group of Heme A". Biochemistry. 43 (27): 8616–8624. doi:10.1021/bi049056m. PMID 15236569.
- Yoshikawa, S. (2012). "Structural studies on bovine heart cytochrome c oxidase". Biochim. Biophys. Acta. 1817 (4): 579–589. doi:10.1016/j.bbabio.2011.12.012. PMID 22236806.
- Rae, T.; Goff, H. (1998). "The heme prosthetic group of lactoperoxidase. Structural characteristics of heme l and heme l-peptides". The Journal of Biological Chemistry. 273 (43): 27968–27977. doi:10.1074/jbc.273.43.27968. PMID 9774411. S2CID 25780396.
- Purdy, M.A. (1983). "Effect of growth phase and cell envelope structure on susceptibility of Salmonella triumphant to the lactoperoxidase-thiocyanate-hydrogen peroxide system". Infect. Immun. 39 (3): 1187–95. doi:10.1128/IAI.39.3.1187-1195.1983. PMC 348082. PMID 6341231.
- Ohshima, H. (2003). "Chemical basis of inflammation-induced carcinogenesis". Arch. Biochem. Biophys. 417 (1): 3–11. doi:10.1016/s0003-9861(03)00283-2. PMID 12921773.
- Henderson, J.P. (2003). "Phagocytes produce 5-chlorouracil and 5-bromouracil, two mutagenic products of myeloperoxidase, in human inflammatory tissue". J. Biol. Chem. 278 (26): 23522–8. doi:10.1074/jbc.m303928200. PMID 12707270. S2CID 19631565.
- Murshudov, G.; Grebenko, A.; Barynin, V.; Dauter, Z.; Wilson, K.; Vainshtein, B.; Melik-Adamyan, W.; Bravo, J.; Ferrán, J.; Ferrer, J. C.; Switala, J.; Loewen, P. C.; Fita, I. (1996). "Structure of the heme d of Penicillium vitale and Escherichia coli catalases" (PDF). The Journal of Biological Chemistry. 271 (15): 8863–8868. doi:10.1074/jbc.271.15.8863. PMID 8621527. Archived (PDF) from the original on 2018-07-24.
- Belevich, I. (2005). "Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability". FEBS Letters. 579 (21): 4567–70. doi:10.1016/j.febslet.2005.07.011. PMID 16087180. S2CID 36465802.
- Fischer, H.; Orth, H. (1934). Die Chemie des Pyrrols. Liepzig: Ischemia Verlagsgesellschaft.
- Puustinen A, Wikström M. (1991). "The heme groups of cytochrome o from Escherichia coli". Proc. Natl. Acad. Sci. U.S.A. 88 (14): 6122–6. Bibcode:1991PNAS...88.6122P. doi:10.1073/pnas.88.14.6122. PMC 52034. PMID 2068092.
- Caughey WS, York JL (1962). "Isolation and some properties of the green heme of cytochrome oxidase from beef heart muscle". J. Biol. Chem. 237 (7): 2414–6. doi:10.1016/S0021-9258(19)63456-3. PMID 13877421.
- Caughey WS, Smythe GA, O'Keeffe DH, Maskasky JE, Smith ML (1975). "Heme A of cytochrome c oxidase. Structure and properties: comparisons with hemes B, C, and S and derivatives". J. Biol. Chem. 250 (19): 7602–22. doi:10.1016/S0021-9258(19)40860-0. PMID 170266.
- Battersby, Alan R. (2000). "Tetrapyrroles: The pigments of life". Natural Product Reports. 17 (6): 507–526. doi:10.1039/B002635M. PMID 11152419.
- Sridevi, Kolluri (28 April 2018). Upregulation of Heme Pathway Enzyme ALA Synthase-1 by Glutethimide and 4,6-Dioxoheptanoic Acid and Downregulation by Glucose and Heme: A Dissertation. EScholarship@UMMS (Thesis). University of Massachusetts Medical School. doi:10.13028/yyrz-qa79. Archived from the original on 8 August 2016. Retrieved 28 April 2018.
- Kumar, Sanjay; Bandyopadhyay, Uday (July 2005). "Free heme toxicity and its detoxification systems in human". Toxicology Letters. 157 (3): 175–188. doi:10.1016/j.toxlet.2005.03.004. PMID 15917143.
- Puy, Hervé; Gouya, Laurent; Deybach, Jean-Charles (March 2010). "Porphyrias". The Lancet. 375 (9718): 924–937. doi:10.1016/S0140-6736(09)61925-5. PMID 20226990. S2CID 208791867.
- Fraser, Rachel Z.; Shitut, Mithila; Agrawal, Puja; Mendes, Odete; Klapholz, Sue (2018-04-11). "Safety Evaluation of Soy Leghemoglobin Protein Preparation Derived FromPichia pastoris, Intended for Use as a Flavor Catalyst in Plant-Based Meat". International Journal of Toxicology. 37 (3): 241–262. doi:10.1177/1091581818766318. ISSN 1091-5818. PMC 5956568. PMID 29642729.
- "Inside the Strange Science of the Fake Meat That 'Bleeds'". Wired. 2017-09-20. Archived from the original on 24 March 2018. Retrieved 28 April 2018.
- "Silicon Valley gets a taste for food". The Economist. 2015-03-05. ISSN 0013-0613. Retrieved 2019-04-08.
- Maines, Mahin D. (July 1988). "Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications". The FASEB Journal. 2 (10): 2557–2568. doi:10.1096/fasebj.2.10.3290025. ISSN 0892-6638. PMID 3290025. S2CID 22652094.
- Lehninger's Principles of Biochemistry (5th ed.). New York: W. H. Freeman and Company. 2008. pp. 876. ISBN 978-0-7167-7108-1.
- Hanafy, K.A. (2013). "Carbon Monoxide and the brain: time to rethink the dogma". Curr. Pharm. Des. 19 (15): 2771–5. doi:10.2174/1381612811319150013. PMC 3672861. PMID 23092321.
- Abraham, N.G.; Kappas, A. (2008). "Pharmacological and clinical aspects of heme oxygenase". Pharmacol. Rev. 60 (1): 79–127. doi:10.1124/pr.107.07104. PMID 18323402. S2CID 12792155.
- Florczyk, U.M.; Jozkowicz, A.; Dulak, J. (January–February 2008). "Biliverdin reductase: new features of an old enzyme and its potential therapeutic significance". Pharmacological Reports. 60 (1): 38–48. PMC 5536200. PMID 18276984.
- King, C.; Rios, G.; Green, M.; Tephly, T. (2000). "UDP-Glucuronosyltransferases". Current Drug Metabolism. 1 (2): 143–161. doi:10.2174/1389200003339171. PMID 11465080.
- Helmenstine, Anne Marie. "The Chemicals Responsible for the Color of Urine and Feces". ThoughtCo. Retrieved 2020-01-24.
- Bunn, H. F.; Jandl, J. H. (Sep 1966). "Exchange of heme among hemoglobin molecules". Proc. Natl. Acad. Sci. USA. 56 (3): 974–978. Bibcode:1966PNAS...56..974B. doi:10.1073/pnas.56.3.974. PMC 219955. PMID 5230192.
- Smith, M. L.; Paul, J.; Ohlsson, P. I.; Hjortsberg, K.; Paul, K. G. (Feb 1991). "Heme-protein fission under nondenaturing conditions". Proc. Natl. Acad. Sci. USA. 88 (3): 882–886. Bibcode:1991PNAS...88..882S. doi:10.1073/pnas.88.3.882. PMC 50918. PMID 1846966.
- Everse, J.; Hsia, N. (1197). "The toxicities of native and modified hemoglobins". Free Radical Biology and Medicine. 22 (6): 1075–1099. doi:10.1016/S0891-5849(96)00499-6. PMID 9034247.
- Kumar, Sanjay; Bandyopadhyay, Uday (July 2005). "Free heme toxicity and its detoxification systems in humans". Toxicology Letters. 157 (3): 175–188. doi:10.1016/j.toxlet.2005.03.004. PMID 15917143.
- Pamplona, A.; Ferreira, A.; Balla, J.; Jeney, V.; Balla, G.; Epiphanio, S.; Chora, A.; Rodrigues, C. D.; Gregoire, I. P.; Cunha-Rodrigues, M.; Portugal, S.; Soares, M. P.; Mota, M. M. (Jun 2007). "Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria". Nature Medicine. 13 (6): 703–710. doi:10.1038/nm1586. PMID 17496899. S2CID 20675040.
- Larsen, R.; Gozzelino, R.; Jeney, V.; Tokaji, L.; Bozza, F. A.; Japiassú, A. M.; Bonaparte, D.; Cavalcante, M. M.; Chora, A.; Ferreira, A.; Marguti, I.; Cardoso, S.; Sepúlveda, N.; Smith, A.; Soares, M. P. (2010). "A central role for free heme in the pathogenesis of severe sepsis". Science Translational Medicine. 2 (51): 51ra71. doi:10.1126/scitranslmed.3001118. PMID 20881280. S2CID 423446.
- Bastide, N. M.; Pierre, F. H.; Corpet, D. E. (2011). "Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved" (PDF). Cancer Prev. Res. 4 (2): 177–184. doi:10.1158/1940-6207.CAPR-10-0113. PMID 21209396. S2CID 4951579. Archived (PDF) from the original on 2015-09-25.
- Bastide, Nadia M.; Pierre, Fabrice H. F.; Corpet, Denis E. (1 February 2011). "Heme Iron from Meat and Risk of Colorectal Cancer: A Meta-analysis and a Review of the Mechanisms Involved". Cancer Prevention Research. 4 (2): 177–184. doi:10.1158/1940-6207.CAPR-10-0113. PMID 21209396. S2CID 4951579. Archived from the original on 11 October 2017. Retrieved 28 April 2018 – via cancerpreventionresearch.aacrjournals.org.
- "Diet, nutrition, physical activity and colorectal cancer". wcrf.org. Retrieved 12 February 2022.
- Chang, Vicky C; Cotterchio, Michelle; Khoo, Edwin (2019). "Iron intake, body iron status, and risk of breast cancer: a systematic review and meta-analysis". BMC Cancer. 19 (1): 543. doi:10.1186/s12885-019-5642-0. PMC 6555759. PMID 31170936.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Plewinska, Magdalena; Thunell, Stig; Holmberg, Lars; Wetmur, James; Desnick, Robert (1991). "delta-Aminolevulinate dehydratase deficient porphyria: identification of the molecular lesions in a severely affected homozygote". American Journal of Human Genetics. 49 (1): 167–174. PMC 1683193. PMID 2063868.
- Aurizi, C.; Lupia Palmieri, G.; Barbieri, L.; Macri, A.; Sorge, F.; Usai, G.; Biolcati, G. (February 2009). "Four novel mutations of the coproporphyrinogen III oxidase gene". Cellular and Molecular Biology. 55 (1): 8–15. PMID 19267996.
- Bustad, H. J.; Vorland, M.; Ronneseth, E.; Sandberg, S.; Martinez, A.; Toska, K. (August 8, 2013). "Conformational stability and activity analysis of two hydroxymethylbilane synthase mutants, K132N and V215E, with different phenotypic association with acute intermittent porphyria". Bioscience Reports. 33 (4): 617–626. doi:10.1042/BSR20130045. PMC 3738108. PMID 23815679.
- Martinez di Montemuros, F.; Di Pierro, E.; Patti, E.; Tavazzi, D.; Danielli, M. G.; Biolcati, G.; Rocchi, E.; Cappllini, M. D. (December 2002). "Molecular characterization of porphyrias in Italy: a diagnostic flow-chart". 48 (8): 867–876.
{{cite journal}}: Cite journal requires|journal=(help) - Badenas, C.; To Figueras, J.; Phillips, J. D.; Warby, C. A.; Muñoz, C.; Herrero, C. (April 2009). "Identification and characterization of novel uroporphyrinogen decarboxylase gene mutations in a large series of porphyria cutanea tarda patients and relatives". Clinical Genetics. 75 (4): 346–353. doi:10.1111/j.1399-0004.2009.01153.x. PMC 3804340. PMID 19419417.