Histone-like nucleoid-structuring protein
Histone-like nucleoid-structuring protein (H-NS), is one of twelve nucleoid-associated proteins (NAPs)[1] whose main function is the organization of genetic material, including the regulation of gene expression via xenogeneic silencing.[2] H-NS is characterized by an N-terminal domain (NTD) consisting of two dimerization sites, a linker region that is unstructured and a C-terminal domain (CTD) that is responsible for DNA-binding.[2] Though it is a small protein (15 kDa),[3] it provides essential nucleoid compaction and regulation of genes (mainly silencing)[2] and is highly expressed, functioning as a dimer or multimer.[3] Change in temperature causes H-NS to be dissociated from the DNA duplex, allowing for transcription by RNA polymerase, and in specific regions lead to pathogenic cascades in enterobacteria such as Escherichia coli and the four Shigella species.[3]

| H-NS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Solution structure of the N-terminal domain (oligomerization domain) of the bacterial nucleoid structuring protein, H-NS. | |||||||||
| Identifiers | |||||||||
| Symbol | H-NS | ||||||||
| Pfam | PF00816 | ||||||||
| InterPro | IPR001801 | ||||||||
| CATH | [ P0ACF8] | ||||||||
| SCOP2 | 1hns / SCOPe / SUPFAM | ||||||||
| |||||||||
Structure
H-NS has a specific topology that allows it to condense bacterial DNA into a superhelical structure based on evidence from X-ray crystallography.[2] The condensed superhelical structure has implicated H-NS in gene repression caused by the formation of oligomers. These oligomers form due to dimerization of two sites in the N-terminal domain of H-NS.[2] For example, in bacterial species like Salmonella typhimurium, the NTD of H-NS contains dimerization sites in helices alpha 1, alpha 2 and alpha 3. Alpha helices 3 and 4 are then responsible for creating the superhelical structure of H-NS-DNA interactions by head to head association (Figure 2).[2][5] H-NS also contains an unstructured linker region, also known as a Q-linker.[2] The C-Terminal domain, also known as the DNA Binding Domain (DBD), shows high affinity for regions in DNA that are rich in Adenine and Thymine and present in a hook-like motif in a minor groove.[2] The base stacking present in this AT rich region of the DNA allows for minor widening of the minor groove that is preferential for binding.[2] Common DBD's include AACTA and TACTA regions which can appear hundreds of times throughout the genome.[2] Within these AT-rich regions, the minor groove has a width of 3.5 Å,[3] which is preferential for H-NS binding. In E. coli, it was observed that H-NS restructures the genome into microdomains in vivo.[2] While the bacterial genome is split into four different macrodomains including Ori and Ter (macrodomain of E. coli and Shigella spp. in which H-NS is encoded),[3] it is thought that H-NS plays a role in the formation of these small 10 kb microdomains throughout the genome.[2]

Function
A major function of H-NS is to influence DNA topology (Figure 2). H-NS is responsible for formation of nucleofilaments along the DNA and DNA-DNA bridges. H-NS is known as a passive DNA bridger, meaning that it binds two distant segments of DNA and remains stationary, forming a loop. This DNA loop formation allows H-NS to control gene expression.[6] Relief of suppression by H-NS can be achieved by the binding of another protein, or by changes in DNA topology which can occur due to changes in temperature and osmolarity, for example.[7] The CTD binds to the bacterial DNA in such a way that inhibits the function of RNA polymerase. This is a common feature seen in horizontally acquired genes.[8] Structural studies of H-NS use bacterial species such as E. coli and Shigella spp. because the C-Terminal Domain is completely conserved.[3]
The process for formation of H-NS-DNA complexes begins with the CTD binding to a preferential site in the genome. This may be the result of the large amount of positively charged amino acid residues located within the linker region that causes the CTD to search for a binding site with high affinity.[2] Once the CTD is bound to its preferential region, TpA step, the NTD's can oligomerize and form rigid nucleofilaments that, if favorable conditions exist, will more freely bind to one another to form DNA-bridges. This form of bridging is known as "passive bridging" and may not allow RNAP to proceed with transcription.[2] The experiments used to support this method of DNA binding and gene silencing come from Atomic Force Microscopy and single-molecule studies in vitro.[2]
All bacteria must be sensitive to changes in their physical environment to survive. These mechanisms allow for turning genes on or off depending on its extracellular environment.[3] Many researchers believe that H-NS contributes to these sensory functions. H-NS has been observed to control around 60% of the temperature regulated genes and can dissociate from the DNA duplex at 37°C.[2] This particular sensitivity seen in H-NS allows for pathogenesis and is the main focus of study. Outside of a host, the temperature of 32°C prevents dissociation of H-NS from the virulence plasmid in Shigella spp. in order to conserve energy for energetically costly production of proteins involved in pathogenesis.[9] The presence of magnesium ions (Mg2+) has been shown to allow H-NS to form a slightly open to completely open conformational change in structure that will ultimately alter the interaction between the negatively charged NTD and positively charged CTD.[2] Magnesium concentrations below 2 mM, allows for the formation of rigid nucleoprotein filaments and high concentrations promote the formation of H-NS DNA bridges.[10] The charges seen in the NTD and CTD may explain how H-NS remains sensitive to changes in temperature and osmolarity (pH below 7.4).[3] H-NS can also interact with other proteins and influence their function, for example it can interact with the flagellar motor protein FliG to increase its activity.[7]
Clinical Significance
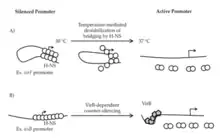
H-NS has a conserved role in the pathogenicity of gram-negative bacteria including Shigella spp., Escherichia coli, Salmonella spp., and many others. It is implicated in the transcription of the virF gene causing what is known as the virF leading to bacillary dysentery, a disease affecting children mainly seen in developing countries. These two bacterial species contain a virulence plasmid that is responsible for invasion of host cells and is regulated by H-NS.[11] Interestingly, almost 70% of the open reading frames (ORF) of the specialized virulence plasmid in Shigella spp. is AT-rich, allowing for long term regulation of this plasmid by H-NS.[3]
Aforementioned, studies show that temperature sensitive H-NS will dissociate from bacterial DNA at 37°C, triggering RNA polymerase to transcribe virF, the gene responsible for the expression of VirF. VirF is the main regulator of the virulence cascade and is expressed due to the temperature sensitive "hinge" region of the virF promoter changing conformation so that is no longer favorable for DNA-bridging by H-NS (Figure 3).[3] Once VirF is expressed, it up regulates the production of icsA, functions to promote motility, and virB, encodes the next regulation protein in the Shigella cascade. As soon as VirB is expressed, it will disrupt H-NS for the rest of the virulence plasmid.[3]
Shigella spp. contain "molecular backups", or paralogues, to H-NS that have been studied in detail due to their apparent assistance in organization of the virulence plasmid.[3] StpA is a paralogue of H-NS that is conserved across the species but the other, Sfh is expressed solely in the S. flexneri mutant strain 2457T.[3] This mutant strain is of much interest to researchers because it acts as a replacement for H-NS since 2457T does not contain the hns gene. The correlation between H-NS and its paralogues is poorly understood at this time.[3] Due to importance of these paralogues in the absence of H-NS in the mutant, further research and focus on these paralogues could lead to promising antibacterial treatments.[3]
References
- Winardhi RS, Yan J, Kenney LJ (October 2015). "H-NS Regulates Gene Expression and Compacts the Nucleoid: Insights from Single-Molecule Experiments". Biophysical Journal. 109 (7): 1321–1329. Bibcode:2015BpJ...109.1321W. doi:10.1016/j.bpj.2015.08.016. PMC 4601063. PMID 26445432.
- Qin L, Erkelens AM, Ben Bdira F, Dame RT (December 2019). "The architects of bacterial DNA bridges: a structurally and functionally conserved family of proteins". Open Biology. 9 (12): 190223. doi:10.1098/rsob.190223. PMC 6936261. PMID 31795918.
- Picker MA, Wing HJ (December 2016). "H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species". Genes. 7 (12): 112. doi:10.3390/genes7120112. PMC 5192488. PMID 27916940.
- Shindo H, Iwaki T, Ieda R, Kurumizaka H, Ueguchi C, Mizuno T, et al. (February 1995). "Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli". FEBS Letters. 360 (2): 125–131. doi:10.1016/0014-5793(95)00079-o. PMID 7875316. S2CID 44479751.
- Bloch V, Yang Y, Margeat E, Chavanieu A, Augé MT, Robert B, et al. (March 2003). "The H-NS dimerization domain defines a new fold contributing to DNA recognition". Nature Structural Biology. 10 (3): 212–218. doi:10.1038/nsb904. PMID 12592399. S2CID 25761309.
- Qin L, Erkelens AM, Ben Bdira F, Dame RT (December 2019). "The architects of bacterial DNA bridges: a structurally and functionally conserved family of proteins". Open Biology. 9 (12): 190223. doi:10.1098/rsob.190223. PMC 6936261. PMID 31795918.
- Donato GM, Kawula TH (September 1998). "Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli". The Journal of Biological Chemistry. 273 (37): 24030–24036. doi:10.1074/jbc.273.37.24030. PMID 9727020.
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC (August 2006). "H-NS mediates the silencing of laterally acquired genes in bacteria". PLOS Pathogens. 2 (8): e81. doi:10.1371/journal.ppat.0020081. PMC 1550270. PMID 16933988.
- Dorman CJ (September 2014). "H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria". Plasmid. 75: 1–11. doi:10.1016/j.plasmid.2014.06.004. PMID 24998344.
- Verma SC, Qian Z, Adhya SL (December 2019). "Architecture of the Escherichia coli nucleoid". PLOS Genetics. 15 (12): e1008456. doi:10.1371/journal.pgen.1008456. PMC 6907758. PMID 31830036.
- Picker MA, Wing HJ (December 2016). "H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species". Genes. 7 (12): E112. doi:10.3390/genes7120112. PMC 5192488. PMID 27916940.