Influenza virus pseudoknot
The Influenza virus pseudoknot is an RNA pseudoknot structure formed in one of the non-structural coding segments (NS) of influenza virus genome.[1] Pseudoknots are commonly found in viral genomes, especially RNA viruses, where they incorporate an RNA splice site and can have a wide range of functions.[2] The orientation of the coaxially stacked stems in the influenza pseudoknot, however, differs from the most common topology in "classical" RNA pseudoknots.[3][4]
| Influenza pseudoknot | |
|---|---|
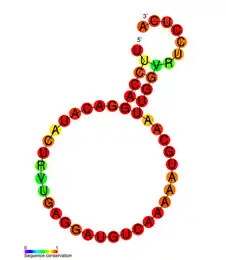 The consensus secondary structure of the Influenza pseudoknot family of RNA motifs | |
| Identifiers | |
| Symbol | PK-IAV |
| Rfam | RF01099 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Orthomyxoviridae; |
| PDB structures | PDBe |
The pseudoknot structure is very similar in influenzavirus A and influenzavirus B. A unique point mutation occurring in the strains of influenza A virus subtype H5N1 after 2001 has been suggested to result in RNA conformational shift,[5] favouring an alternative hairpin structure instead of the pseudoknot.[1]
Another pseudoknot occurs at the Influenza A Segment 7 Splice Site, which is used to produce the important viral M2 ion channel protein.[6][7] Both pseudoknots have the possibility of alternating between hairpin loop and pseudoknot conformations, which place splicing regulatory motifs in different structural contexts.
See also
References
- Gultyaev AP, Olsthoorn RC (March 2010). "A family of non-classical pseudoknots in influenza A and B viruses". RNA Biol. 7 (2): 125–129. doi:10.4161/rna.7.2.11287. PMID 20200490. Retrieved 2010-07-13.
- Brierley I, Pennell S, Gilbert RJ (August 2007). "Viral RNA pseudoknots: versatile motifs in gene expression and replication". Nat. Rev. Microbiol. 5 (8): 598–610. doi:10.1038/nrmicro1704. PMC 7096944. PMID 17632571.
- Pleij CW (April 1990). "Pseudoknots: a new motif in the RNA game". Trends Biochem. Sci. 15 (4): 143–147. doi:10.1016/0968-0004(90)90214-v. PMID 1692647.
- Pleij, C. (1994). "RNA pseudoknots". Current Opinion in Structural Biology. 4 (3): 337–344. doi:10.1016/S0959-440X(94)90101-5. PMC 7133496.
- Gultyaev AP, Heus HA, Olsthoorn RC (February 2007). "An RNA conformational shift in recent H5N1 influenza A viruses". Bioinformatics. 23 (3): 272–276. doi:10.1093/bioinformatics/btl559. PMID 17090581.
- Moss WN, Dela-Moss LI, Kierzek E, Kierzek R, Priore SF, Turner DH (March 2012). Lewin A (ed.). "The 3′ Splice Site of Influenza A Segment 7 mRNA Can Exist in Two Conformations: A Pseudoknot and a Hairpin". PLOS ONE. 7 (6): e38323. Bibcode:2012PLoSO...738323M. doi:10.1371/journal.pone.0038323. PMC 3369869. PMID 22685560.
- Moss WN, Dela-Moss LI, Priore SF, Turner DH (October 2012). "The influenza A segment 7 mRNA 3' splice site pseudoknot/hairpin family". RNA Biology. 9 (11): 1305–1310. doi:10.4161/rna.22343. PMC 3597570. PMID 23064116. Archived from the original on 2013-10-13.