Informative site
In phylogenetics, informative site is a term used when maximum parsimony is the optimality criterion for construction of a phylogenetic tree. It refers to a characteristic for which the number of character-state evolutionary changes of at this site depends on the topology of the tree.[1] The charactetistics can take on multiple types of data, including morphological (such as the presence of wings, tentacles, etc.) or molecular information such as sequences of DNA or proteins.
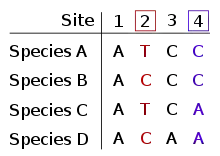
The informative site is a position in the relevant set of aligned sequences at which there are at least two different character states and each of those states occurs in at least two of the sequences. In other words, it cannot be a fully conserved (i.e., invariable) site nor can it be a (singleton) site with a difference in only one sequence (as seen, for example, in single-nucleotide polymorphisms and single-nucleotide variants). In both cases, the number of character-state changes is the same regardless of the topology of the tree, equal to 0 and 1 respectively.[1]
References
- Zvelebil, Marketa J. (2008). Understanding bioinformatics. New York: Garland Science. ISBN 978-0-8153-4024-9.