L-Tryptophan decarboxylase
L-Tryptophan decarboxylase (EC 4.1.1.105) is an enzyme distinguished by the substrate L-tryptophan.[2][3]
| alcohol dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
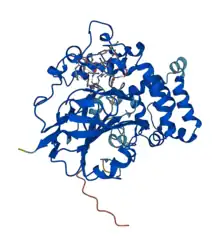 Crystal structure prediction for L--tryptophan decarboxylase .[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.1.105 | ||||||||
| Alt. names | aldehyde reductase | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
This enzyme catalyzes the reaction of L-tryptophan to tryptamine and carbon dioxide.[2][4] The enzymatic reaction namely takes place in the species Psilocybe cubensis, where a decarboxylase, kinase, and methyltransferase work together to synthesize psilocybin.[5][6]
Classification
The enzyme commission number for L-tryptophan decarboxylase is EC 4.1.1.105.[2] Other common names include psilocybin biosynthesis decarboxylase and psiD.[4] The first digit in the enzyme number is representative of the class of enzymes known as lyases, which catalyze elimination reactions.[2][4] The second and third digits are representative of the subclass of lyases known as decarboxylases that cleave carbon-carbon bonds.[2][4] The last digit is representative of the enzyme’s specific substrate, L-tryptophan.[2]
This enzyme is a part of the PLP-independent phosphatidylserine decarboxylase family and most compared to hypothetical proteins of other basidiomycetes fungi.[5] These include Fibulorhizoctonia sp with 60% identical amino acids and Moniliophthora roreri with 52% identical amino acids.[5] A similar enzyme that is not related to L-tryptophan decarboxylase is called aromatic-L-amino-acid decarboxylase with an enzyme number of EC 4.1.1.28.[2]
Reaction pathway
The first step in the reaction is the substrate binding of L-tryptophan, which reacts with a coenzyme hydrogen.[2] The decarboxylase enzyme is able to transform L-tryptophan to tryptamine in the second step by cleaving off two oxygens and a carbon to form tryptamine and carbon dioxide as the products.[2] Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, which makes the molecule polar, while tryptamines have an indole ring structure, a fused double ring consisting of a pyrrole ring, and a benzene ring, which is joined to an amino group by two carbon side chains.[7]
This is the chemical reaction that takes place:
- H+ + C11H12N2O2 = CO2 + C10H13N2
Organisms in which L-tryptophan decarboxylase is found
L-Tryptophan decarboxylase has been characterized in bacteria, plants, and fungi.[2] Fungi that produce psilocybin and psilocin express incredible diversity, as they are a part of at least eight genera with hundreds of species belonging to them.[3] The specific reaction pathway for L-tryptophan decarboxylase has been described in twelve species, including Psilocybe cubensis, commonly known as magic mushrooms.[2] All fungi in the genus Psilocybe have a well-defined, umbrella-like cap with gills underneath and a stipe.[3] Other main characteristics of Psilocybe species include purple-brown reproductive spores, the presence of an annulus, and blue bruising with contact.[3] All Psilocybe species are described to feed on microscopic detritus and are found on a variety of surfaces, such as herbivore dung, grasses, roots, wood, and soil.[3] Humans have a documented history of ingesting this psilocybin producing fungi.[3] There are 57 species that are found in Mexico, and out of these there have been reports of 35 species and nine varieties being used by ethnic groups.[3]
Function
In Psilocybe cubensis, L-tryptophan decarboxylase has been described with two other enzymes to biosynthesize psilocybin in a one pot reaction.[4][5] These other two enzymes in this process are psiK, an enzyme that catalyzes the phosphotransfer step, and psiM, an enzyme that catalyzes the iterative N-methyl transfer step.[4] The biosynthesis of psilocybin takes place as follows:
L-Tryptophan is decarboxylated to 4-hydroxytryptamine by psiD → psiK phosphorylates 4-hydroxytryptamine to create norbaeocystin → psiM then processively N,N-dimethylates the compound to yield psilocybin.[4]
Serotonin 2A receptors (5-HT2ARs) stimulation by the active metabolite, psilocin, disrupts serotonergic neurotransmission and produces the characteristic psychedelic effects of this species of fungus.[6] The product formed by L-tryptophan decarboxylase, tryptamine, is relevant to humans because the mammalian brain contains very low concentrations of tryptamine; and serotonin is a tryptamine natural derivative involved in regulating central nervous system processes like sleep, cognition, memory, temperature regulation and behavior.[7] In contrast, the naturally occurring derivatives of tryptamine are found in magic mushrooms (Psilocybe cubensis).[7]
Structure
L-Tryptophan decarboxylase is 439 amino acid residues long in its native form and a calculated pI 5.3.[4] The crystal structure of L-tryptophan decarboxylase has been modeled and predicted by AlphaFold with an average confidence of 91.17% and SWISS-MODEL with an average confidence of 25.37% as an oligo-state monomer, but the crystal structure remains to be described.[1][8]
Active sites
4-Hydroxy-L-tryptophan is accepted as a substrate by the enzyme in addition to L-tryptophan.[4][5] This subsequent pathway is suggested to yield 4-Hydroxytryptamine instead of tryptamine.[4] Both of these compounds can be used in the biosynthesis of psilocybin.[4] The enzyme is distinct from other fungal and plant aromatic amino acid decarboxylases because it belongs to a class that L-tryptophan has not previously been described as a substrate for.[4] Currently, the active sites for L-tryptophan decarboxylase remain to be described.
Evolution
Due to the multi-step process of psilocybin biosynthesis and its restricted phylogenetic distribution, the pathway involving L-tryptophan decarboxylase has been suggested to evolve via horizontal gene cluster transfer.[9][10] The phylogenies of the genes involved in psilocybin biosynthesis (including L-tryptophan decarboxylase) suggests that the process first evolved in wood-decaying fungi, and then evolved in dung-decaying fungi through vertical and horizontal gene transfer due to shared environmental pressures.[9][10] Neurological effects from psilocybin in wood and dung decaying fungi posit that psilocybin could be an ecological modulator acting on insect behavior.[9] This would advantage the fungi by disrupting and inhibiting the behavior of competitors, such as termites and fruit fly larvae, for wood and dung resources.[9][11] Invertebrate competitors that would be especially impacted by this are social insects, where neurotransmitter mimics would disrupt coordination.[12]
References
- "L-tryptophan decarboxylase". AlphaFold Protein Structure Database. Retrieved 2022-10-21.
- "Information on EC 4.1.1.105 - L-tryptophan decarboxylase". BRENDA Enzyme Database. Retrieved 2022-10-20.
- Van Court RC, Wiseman MS, Meyer KW, Ballhorn DJ, Amses KR, Slot JC, et al. (April 2022). "Diversity, biology, and history of psilocybin-containing fungi: Suggestions for research and technological development". Fungal Biology. 126 (4): 308–319. doi:10.1016/j.funbio.2022.01.003. PMID 35314062. S2CID 246793389.
- Fricke J, Blei F, Hoffmeister D (September 2017). "Enzymatic Synthesis of Psilocybin". Angewandte Chemie. 56 (40): 12352–12355. doi:10.1002/anie.201705489. PMID 28763571.
- Blei F, Baldeweg F, Fricke J, Hoffmeister D (May 2018). "Biocatalytic Production of Psilocybin and Derivatives in Tryptophan Synthase-Enhanced Reactions". Chemistry: A European Journal. 24 (40): 10028–10031. doi:10.1002/chem.201801047. PMID 29750381. S2CID 13702079.
- Madsen MK, Fisher PM, Burmester D, Dyssegaard A, Stenbæk DS, Kristiansen S, et al. (June 2019). "Psychedelic effects of psilocybin correlate with serotonin 2A receptor occupancy and plasma psilocin levels". Neuropsychopharmacology. 44 (7): 1328–1334. doi:10.1038/s41386-019-0324-9. PMC 6785028. PMID 30685771.
- Tittarelli R, Mannocchi G, Pantano F, Romolo FS (January 2015). "Recreational use, analysis and toxicity of tryptamines". Current Neuropharmacology. 13 (1): 26–46. doi:10.2174/1570159X13666141210222409. PMC 4462041. PMID 26074742.
- "L-tryptophan decarboxylase". SWISS-MODEL Repository. Expasy, Swiss Institute of Bioinformatics (SIB). P0DPA6. Retrieved 2022-10-21.
- Reynolds HT, Vijayakumar V, Gluck-Thaler E, Korotkin HB, Matheny PB, Slot JC (April 2018). "Horizontal gene cluster transfer increased hallucinogenic mushroom diversity". Evolution Letters. 2 (2): 88–101. doi:10.1002/evl3.42. PMC 6121855. PMID 30283667.
- Awan AR, Winter JM, Turner D, Shaw WM, Suz LM, Bradshaw AJ, Ellis T, Dentinger BT (2018-07-27). "Convergent evolution of psilocybin biosynthesis by psychedelic mushrooms". bioRxiv: 374199. doi:10.1101/374199. S2CID 91735116.
- Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, et al. (December 2007). "A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation". Science. 318 (5858): 1913–1916. Bibcode:2007Sci...318.1913H. doi:10.1126/science.1146954. PMID 18096805. S2CID 19392955.
- Genise JF (2017). "The Trace Fossil Record of Eusociality in Ants and Termites". In Genise JF (ed.). Ichnoentomology: Insect Traces in Soils and Paleosols. Topics in Geobiology. Vol. 37. Cham: Springer International Publishing. pp. 285–312. doi:10.1007/978-3-319-28210-7_12. ISBN 978-3-319-28210-7.