MFSD6L
Major facilitator superfamily domain containing 6 like (MFSD6L) is a protein encoded by the MFSD6L gene in humans.[5] The MFSD6L protein is a transmembrane protein that is part of the major facilitator superfamily (MFS) that uses chemiosmotic gradients to facilitate the transport of small solutes across cell membranes.
| MFSD6L | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MFSD6L, major facilitator superfamily domain containing 6 like | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 2384904 HomoloGene: 17128 GeneCards: MFSD6L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
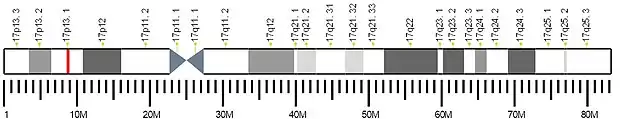
In the human genome, the MFSD6L gene is located on chromosome 17 (17p13.1).[5] The DNA sequence encoding the polypeptide encompasses 2,256 bases, starting from 8,797,110 bp to 8,799,365 bp.[6] Additionally, the gene sequence resides on the minus strand.[5] The MFSD6L gene has one alias called FLJ35773.[5]
The encoding DNA sequence results in only one exon in the translated mRNA sequence.[6]
The tumor suppressor gene TP53 was also found within the gene neighborhood of MFSD6L at 17p13.1.[7]
mRNA Transcript
The MFSD6L gene was not found to have other isoforms due to the presence of only one exon in the MFSD6L encoding sequence.
Protein
The MFSD6L protein has a precursor molecular weight of approximately 64 kDa, consisting of 586 amino acids.[5] After post-translational modifications, such as glycosylation, the mature MFSD6L protein's molecular weight increases to 72 kDa. Of the amino acids consisting the MFSD6L protein, leucine was found to have increased levels compared to most other human proteins. This increase in leucine is also present in the MFSD6L protein of the house mouse and chimpanzee.[8] The protein also has an isoelectric point of 8.87 pI.[9]
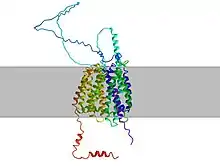
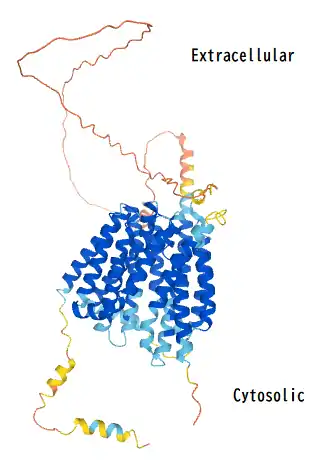
The peptide sequence contains 11 transmembrane regions that cross the plasma membrane. Additionally, there are also two MFS regions starting at the 28th and 368th encoding amino acids.[10]
For the secondary structure of the MFSD6L protein, there are 16 predicted alpha helices and 3 predicted beta sheets.[11] The large amount of alpha helices within the structure of MFSD6L can be attributed to the protein being a transmembrane solute transporter since alpha helices are usually the part of the protein's structure that is positioned within the cell membrane.
Within the tertiary structure, there was a disulfide bond predicted between the two cysteines at the 29th and 311th amino acids.[12]
Expression and regulation
Gene level

There was only one promoter region, spanning 1,107 bp, found for MFSD6L using the Genomatix Gene2Promoter database.[13] For the part of the promoter region closest to the start of the 5' UTR of the MFSD6L gene, there were several transcription factor binding sites found. A transcription factor binding site of note was the site for the p53 tumor suppressor protein.[13]
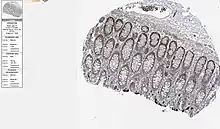
The MFSD6L gene was found to be highly expressed in the pancreas, salivary glands, and the thyroid.[14]
Inspection of in-situ hybridization expression of MFSD6L gene shows that the gene is particularly expressed within glandular cells within their respective tissues.
Expression of the MFSD6L was found to be upregulated as a result of glucose starvation.[15]
Transcript level
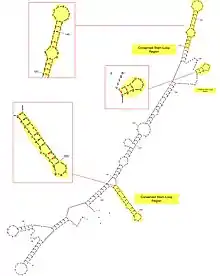
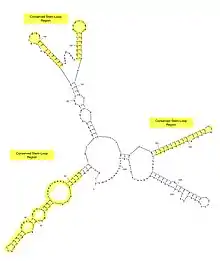
Since there is only one exon and no introns within the MFSD6L gene, There is no splicing performed on the MFSD6L mRNA.[10] Translation of the MFSD6L protein initiates at the end of the 5' UTR, which is the first 245 nucleotides of the MFSD6L mRNA. There are conserved stem-loop regions across mammalian orthologs, which infer possible miRNA binding sites.
Protein level
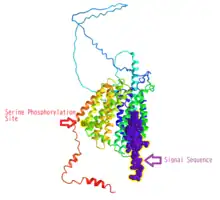
The subcellular localization of the MFSD6L protein is predicted to be within the cell membrane via DeepLoc tool.[16] This is supported by it being a solute symporter similar to MFS proteins. The first 28 amino acids of the translated MFSD6L protein contains the signal peptide.[17]
Additionally, n-glycosylation sites were predicted at the 110th, 129th, and 224th amino acids of the protein sequence.[16] A serine phosphorylation site at the 429th amino acid was also predicted and verified by presence within other mammalian orthologs.[18]
Evolution
Paralogs
The MFSD6 protein was found to be the only paralog to the human MFSD6L protein.[5]
Orthologs
Through BLAST sequence analysis, the MFSD6L protein was found to have orthologs in a many mammalian species, especially among primates, and flying foxes.[19] There were some orthologs found in the Reptilia and Amphibia classes, albeit not as great in number as in the Mammalia. Among fish, there were significantly more orthologs found amongst ray-finned fishes than cartilaginous fishes. Additionally, the jaw-less fish, the sea lamprey, was also found to be an ortholog.
There were also multiple orthologs found amongst invertebrates, such as echinoderms and mollusks.
No significant orthologs of the MFSD6L protein were found amongst insects; however there were orthologs found in the bacteria. Specifically, the Anaerolinea genus, which contains thermophilic bacteria were found to have orthologs with the human protein due to its regions of MFS being identical to MFS regions found in the human protein. The following table shows some examples of orthologs of the human MFSD6L.
| Genus and Species | Common Name | Taxonomic Group | Median Time since Divergence (MYA) | Accession Number (from NCBI) | Sequence
Length (aa) |
Sequence
Identity (%) |
Sequence
Similarity (%) |
| Homo sapiens | Human | Primates | 0 | NP_689812.3 | 586 | 100% | 100% |
| Mus musculus | House Mouse | Rodentia | 89 | NP_666116.1 | 586 | 68% | 77% |
| Monodon monoceros | Narwhal | Artiodactyla | 94 | XP_029068193.1 | 595 | 73% | 81% |
| Molossus molossus | Velvety Free-tailed Bat | Chiroptera | 94 | XP_036125248.1 | 600 | 72% | 79% |
| Dermochelys coriacea | Leatherback Sea Turtle | Testudines | 318 | XP_038228264.1 | 653 | 45% | 59% |
| Terrapene carolina triunguis | Three-toed Box Turtle | Testudines | 318 | XP_024071382.1 | 654 | 44% | 57% |
| Patagioenas fasciata monilis | Band-tailed Pigeon | Columbiformes | 318 | OPJ90083.1 | 655 | 43% | 55% |
| Dromaius novaehollandiae | Emu | Casuariiformes | 318 | XP_025976398.1 | 628 | 42% | 55% |
| Microcaecilia unicolor | Tiny Cayenne Caecilian | Gymnophiona | 351.7 | XP_030063970.1 | 652 | 44% | 58% |
| Xenopus tropicalis | Western-clawed Frog | Anura | 351.7 | XP_002937042.2 | 611 | 42% | 56% |
| Bufo bufo | Common Toad | Anura | 351.7 | XP_040293432.1 | 615 | 39% | 54% |
| Scleropages formosus | Asian Arowana | Osteoglossiformes | 433 | NP_001003586.1 | 585 | 40% | 55% |
| Danio rerio | Zebrafish | Cypriniformes | 433 | XP_018612492.2 | 542 | 33% | 49% |
| Callorhinchus milii | Australian Ghostfish | Chimaeriformes | 465 | XP_042198386.1 | 630 | 41% | 57% |
| Petromyzon marinus | Sea Lamprey | Petromyzontiformes | 599 | XP_032823230.1 | 735 | 26% | 38% |
| Anneissia japonica | Sea Lily | Comatulida | 627 | XP_033125701.1 | 616 | 28% | 47% |
| Gigantopelta aegis | Deep Sea Snail | Neomphalida | 736 | XP_041362795.1 | 637 | 27% | 46% |
| Crassostrea gigas | Pacific Oyster | Ostreida | 736 | XP_011445242.2 | 615 | 25% | 44% |
| Octopus sinesis | East Asian Common Octopus | Octopoda | 736 | XP_029655326 | 634 | 24% | 43% |
| Tetranychus urticae | Red Spider Mite | Trombidiformes | 736 | XP_015795313.1 | 872 | 14% | 25% |
| Anaerolinealis | Anaerolinealis bacterium | Anaerolineales | 4090 | MBN1451601.1 | 389 | 19% | 34% |
Homologous gomains
The main homologous domains found within the MFSD6L protein are the MFS regions. Since MFS includes a large amount of solute transporter proteins within its superfamily, there are many MFS proteins that have the same homologous MFS domains.
Most distant homologs
Through BLAST sequence analysis, the most distant homologs were the organisms within the Cnidaria phylum, which mainly consists of jellyfish, sea anemones, and corals.[19] Searching with BLAST for the MFSD6L gene at an older diverging phylum, the Porifera, revealed no homologous MFSD6L protein.
Predicted emergence date
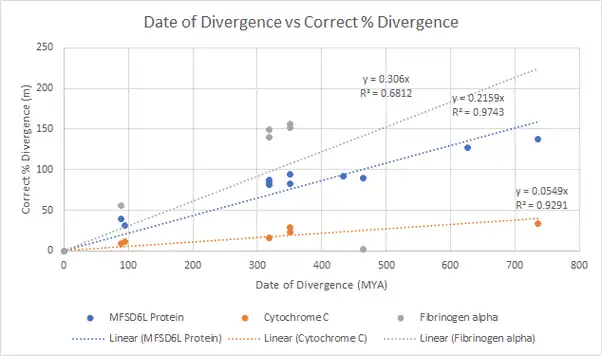
As a result of the MFSD6L protein's presence in Cnidaria and absence in Porifera, the estimated emergence date of the MFSD6L gene lies between 687 and 777 MYA, which are the divergence dates found from TimeTree.[20] From the corrected % divergence chart and calculations of the corrected % divergence of the Homo sapiens MFSD6 paralog, the estimated date of emergence of the MFSD6L protein was found to be around 736 MYA.
Interacting proteins
The MFSD6L protein was not found to have any experimentally-verified protein-protein interactions.[21]
Function
The polypeptide sequence contains many transmembrane regions, identifying the MFSD6L protein as a transmembrane protein for transporting solutes across the plasma membrane of a cell. Tertiary structure prediction tools suggest that the structure of the MFSD6L protein is similar to 1PV6A, a β-galactosides symporter which uses proton gradients to transport solutes.[22][23] As a result, the function of the MFSD6L protein could possibly a sugar symporter. This is additionally supported by the fact that the expression of MFSD6L was upregulated due to glucose starvation.
Clinical significance
Disease association
A disease associated with the MFSD6L gene is the Tetralogy of Fallot, which is a series of four congenital heart defects that can cause low oxygenation of blood.[5] This is due to a ventricular septal defect that causes the mixing of oxygenated and deoxygenated blood in the left ventricle of the heart.
The MFSD6L gene was also found to be a candidate gene taking part in the disease Pediatric Cataract.[24]
Mutations
Various SNP's were found within the encoding sequence of the MFSD6L protein sequence as shown below.
| Amino Acid Position | mRNA Position | Original Nucleotide | SNP | Original Amino Acid | Variant Amino Acid | Codon |
| 328 | 1212 | T | T | Ser [S] | Leu [L] | 2 |
| 399 | 1424 | G | A | Gly [G] | Ser [S] | 1 |
| 406 | 1446 | C | T | Ser [S] | Leu [L] | 2 |
| 571 | 1941 | C | T | Thr [T] | Ile [I] | 2 |
| 574 | 1949 | G | A | Asp [D] | Asn [N] | 1 |
| 575 | 1952 | T | G | Trp [W] | Gly [G] | 1 |
References
- GRCh38: Ensembl release 89: ENSG00000185156 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000048329 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "MFSD6L Gene". www.genecards.org. Retrieved 2021-10-01.
- "MFSD6L major facilitator superfamily domain containing 6 like [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-10-24.
- "TP53 Gene". www.genecards.org. Retrieved 2021-12-15.
- "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2021-12-14.
- "ExPASy - Compute pI/Mw tool". web.expasy.org. Retrieved 2021-12-14.
- "Homo sapiens major facilitator superfamily domain containing 6 like (MFSD6L), mRNA". 2021-06-27.
{{cite journal}}: Cite journal requires|journal=(help) - "PredictProtein - Protein Sequence Analysis, Prediction of Structural and Functional Features". predictprotein.org. Retrieved 2021-12-14.
- "DiANNA". clavius.bc.edu. Retrieved 2021-12-14.
- "Genomatix Software Suite". Genomatix.
- "GDS3113 / 224438". www.ncbi.nlm.nih.gov. Retrieved 2021-12-15.
- Weldai, Lydia (2018-04-16). "Do Major Facilitator Superfamily Domain Containing Proteins Respond to Glucose Starvation?". Digitala Vetenskapliga Arkivet.
- "Services". www.healthtech.dtu.dk. Retrieved 2021-12-16.
- "Protter - interactive protein feature visualization". wlab.ethz.ch. Retrieved 2021-12-16.
- "PhosphoSitePlus". www.phosphosite.org. Retrieved 2021-12-16.
- "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2021-12-17.
- "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 2021-12-17.
- "MFSD6L protein (human) - STRING interaction network". string-db.org. Retrieved 2021-12-17.
- "I-TASSER server for protein structure and function prediction". zhanggroup.org. Retrieved 2021-12-17.
- "lacY - Lactose permease - Escherichia coli (strain K12) - lacY gene & protein". www.uniprot.org. Retrieved 2021-12-17.
- Aldahmesh, Mohammed A.; Khan, Arif O.; Mohamed, Jawahir Y.; Hijazi, Hadia; Al-Owain, Mohammed; Alswaid, Abdulrahman; Alkuraya, Fowzan S. (December 2012). "Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes". Genetics in Medicine. 14 (12): 955–962. doi:10.1038/gim.2012.86. ISSN 1530-0366. PMID 22935719. S2CID 45088616.