MHC restriction
MHC-restricted antigen recognition, or MHC restriction, refers to the fact that a T cell can interact with a self-major histocompatibility complex molecule and a foreign peptide bound to it, but will only respond to the antigen when it is bound to a particular MHC molecule.[1]
When foreign proteins enter a cell, they are broken into smaller pieces called peptides. These peptides, also known as antigens, can derive from pathogens such as viruses or intracellular bacteria. Foreign peptides are brought to the surface of the cell and presented to T cells by proteins called the major histocompatibility complex (MHC). During T cell development, T cells go through a selection process in the thymus to ensure that the T cell receptor (TCR) will not recognize MHC molecule presenting self-antigens, i.e that its affinity is not too high. High affinity means it will be autoreactive, but no affinity means it will not bind strongly enough to the MHC. The selection process results in developed T cells with specific TCRs that might only respond to certain MHC molecules but not others. The fact that the TCR will recognize only some MHC molecules but not others contributes to "MHC restriction". The biological reason of MHC restriction is to prevent supernumerary wandering lymphocytes generation, hence energy saving and economy of cell-building materials. [2]
T-cells are a type of lymphocyte that is significant in the immune system to activate other immune cells. T-cells will recognize foreign peptides through T-cell receptors (TCRs) on the surface of the T cells, and then perform different roles depending on the type of T cell they are in order to defend the host from the foreign peptide, which may have come from pathogens like bacteria, viruses or parasites. Enforcing the restriction that T cells are activated by peptide antigens only when the antigens are bound to self-MHC molecules, MHC restriction adds another dimension to the specificity of T cell receptors so that an antigen is recognized only as peptide-MHC complexes.[3]
MHC restriction in T cells occurs during their development in the thymus, specifically positive selection.[4] Only the thymocytes (developing T cells in the thymus) that are capable of binding, with an appropriate affinity, with the MHC molecules can receive a survival signal and go on to the next level of selection. MHC restriction is significant for T cells to function properly when it leaves the thymus because it allows T cell receptors to bind to MHC and detect cells that are infected by intracellular pathogens, viral proteins and bearing genetic defects. Two models explaining how restriction arose are the germline model and the selection model.
The germline model suggests that MHC restriction is a result of evolutionary pressure favoring T cell receptors that are capable of binding to MHC.[5] The selection model suggests that not all T cell receptors show MHC restriction, however only the T cell receptors with MHC restriction are expressed after thymus selection.[6] In fact, both hypotheses are reflected in the determination of TCR restriction, such that both germline-encoded interactions between TCR and MHC and co-receptor interactions with CD4 or CD8 to signal T cell maturation occur during selection.[7]
Introduction
The TCRs of T cells recognize linear peptide antigens only if coupled with a MHC molecule. In other words, the ligands of TCRs are specific peptide-MHC complexes.[8] MHC restriction is particularly important for self-tolerance, which makes sure that the immune system does not target self-antigens. When primary lymphocytes are developing and differentiating in the thymus or bone marrow, T cells die by apoptosis if they express high affinity for self-antigens presented by an MHC molecule or express too low an affinity for self MHC.[9]
T cell maturation involves two distinct developmental stages: positive selection and negative selection. Positive selection ensures that any T-cells with a high enough affinity for MHC bound peptide survive and goes on to negative selection, while negative selection induces death in T-cells which bind self-peptide-MHC complex too strongly. Ultimately, the T-cells differentiate and mature to become either T helper cells or T cytotoxic cells. At this point the T cells leave the primary lymphoid organ and enter the blood stream.[10]
The interaction between TCRs and peptide-MHC complex is significant in maintaining the immune system against foreign antigens. MHC restriction allows TCRs to detect host cells that are infected by pathogens, contains non-self proteins or bears foreign DNA. However, MHC restriction is also responsible for chronic autoimmune diseases and hypersensitivity.[8]
Structural specificity
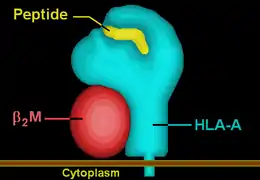
The peptide-MHC complex presents a surface that looks like an altered self to the TCR.[11] The surface consisting of two α helices from the MHC and a bound peptide sequence is projected away from the host cell to the T cells, whose TCRs are projected away from the T cells towards the host cells. In contrast with T cell receptors which recognize linear peptide epitopes, B cell receptors recognize a variety of conformational epitopes (including peptide, carbohydrate, lipid and DNA) with specific three-dimensional structures.[8]
Imposition
The imposition of MHC restriction on the highly variable TCR has caused heated debate. Two models have been proposed to explain the imposition of MHC restriction. The Germline model proposes that MHC restriction is hard-wired in the TCR Germline sequence due to co-evolution of TCR and MHC to interact with each other. The Selection model suggests that MHC restriction is not a hard-wired property in the Germline sequences of TCRs, but imposed on them by CD4 and CD8 co-receptors during positive selection. The relative importance of the two models are not yet determined.[12]
Germline model
The Germline hypothesis suggests that the ability to bind to MHC is intrinsic and encoded within the germline DNA that are coding for TCRs. This is because of evolutionary pressure selects for TCRs that are capable of binding to MHC and selects against those that are not capable of binding to MHC.[13] Since the emergence of TCR and MHC ~500 million years ago,[14] there is ample opportunity for TCR and MHC to coevolve to recognize each other. Therefore, it is proposed that evolutionary pressure would lead to conserved amino acid sequences at regions of contact with MHCs on TCRs.[12]
Evidence from X-ray crystallography has shown comparable binding topologies between various TCR and MHC-peptide complexes.[15] In addition, conserved interactions between TCR and specific MHCs support the hypothesis that MHC restriction is related to the co-evolution of TCR and MHC to some extent.[16]
Selection model
The selection hypothesis argues that instead of being an intrinsic property, MHC restriction is imposed on the T cells during positive thymic selection after random TCRs are produced.[17] According to this model, T cells are capable of recognizing a variety of peptide epitopes independent of MHC molecules before undergoing thymic selection. During thymic selection, only the T cells with affinity to MHC are signaled to survive after the CD4 or CD8 co-receptors also bind to the MHC molecule. This is called positive selection.[18]
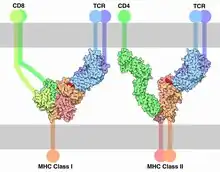
During positive selection, co-receptors CD4 and CD8 initiate a signaling cascade following MHC binding.[19] This involves the recruitment of Lck, a tyrosine kinase essential for T cell maturation that is associated with the cytoplasmic tail of the CD4 or CD8 co-receptors. Selection model argues that Lck is directed to TCRs by co-receptors CD4 and CD8 when they recognize MHC molecules.[4] Since TCRs interact better with Lck when they are binding to the MHC molecules that are binding to the co-receptors in a ternary complex, T cells that can interact with MHCs bound to by the co-receptors can activate the Lck kinase and receive a survival signal.[12]
Supporting this argument, genetically modified T cells without CD4 and CD8 co-receptors express MHC-independent TCRs.[18] It follows that MHC restriction is imposed by CD4 and CD8 co-receptors during positive selection of T cell selection.
Reconciliation
A reconciliation of the two models was offered later on[7] suggesting that both co-receptor and germline predisposition to MHC binding play significant roles in imposing MHC restriction. Since only those T cells that are capable of binding to MHCs are selected for during positive selection in the thymus, to some extent evolutionary pressure selects for germline TCR sequences that bind MHC molecules. On the other hand, as suggested by the selection model, T cell maturation requires the TCRs to bind to the same MHC molecules as the CD4 or CD8 co-receptor during T cell selection, thus imposing MHC restriction.[12]
References
- Immunobiology: The Immune System in Health and Disease. 5th edition. Janeway CA Jr, Travers P, Walport M, et al. New York: Garland Science; 2001. https://www.ncbi.nlm.nih.gov/books/NBK10757/
- Nesmiyanov, P (2020). "Antigen Presentation and Major Histocompatibility Complex". Reference Module in Biomedical Sciences: 90–98. doi:10.1016/B978-0-12-818731-9.00029-X. ISBN 9780128012383. S2CID 234948691.
- Charles A Janeway, Jr; Travers, Paul; Walport, Mark; Shlomchik, Mark J. (2001-01-01). "Antigen Recognition by B-cell and T-cell Receptors". Garland Science.
{{cite journal}}: Cite journal requires|journal=(help) - Van Laethem, François; Tikhonova, Anastasia N.; Singer, Alfred (2012-01-09). "MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection". Trends in Immunology. 33 (9): 437–441. doi:10.1016/j.it.2012.05.006. ISSN 1471-4906. PMC 3427466. PMID 22771139.
- Christopher Garcia, K; Adams, Jarrett J; Feng, Dan; Ely, Lauren K (2009-01-16). "The molecular basis of TCR germline bias for MHC is surprisingly simple". Nature Immunology. 10 (2): 143–147. doi:10.1038/ni.f.219. PMC 3982143. PMID 19148199.
- Collins, Edward J.; Riddle, David S. (2008-08-26). "TCR-MHC docking orientation: natural selection, or thymic selection?". Immunologic Research. 41 (3): 267–294. doi:10.1007/s12026-008-8040-2. ISSN 0257-277X. PMID 18726714. S2CID 33347746.
- Garcia, K. Christopher (2012-09-01). "Reconciling views on T cell receptor germline bias for MHC". Trends in Immunology. 33 (9): 429–436. doi:10.1016/j.it.2012.05.005. PMC 3983780. PMID 22771140.
- Parham, Peter (2005-01-01). "Putting a face to MHC restriction". Journal of Immunology. 174 (1): 3–5. doi:10.4049/jimmunol.174.1.3. ISSN 0022-1767. PMID 15611221.
- B Adkins; C Mueller; C Y Okada; R A Reichert; I L Weissman; Spangrude, G. J. (1987-01-01). "Early Events in T-Cell Maturation". Annual Review of Immunology. 5 (1): 325–365. doi:10.1146/annurev.iy.05.040187.001545. PMID 3109456.
- Klein, Ludger; Kyewski, Bruno; Allen, Paul M.; Hogquist, Kristin A. (2014). "Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see)". Nature Reviews Immunology. 14 (6): 377–391. doi:10.1038/nri3667. PMC 4757912. PMID 24830344.
- Bjorkman, P. J.; Saper, M. A.; Samraoui, B.; Bennett, W. S.; Strominger, J. L.; Wiley, D. C. (1987-10-08). "The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens". Nature. 329 (6139): 512–518. Bibcode:1987Natur.329..512B. doi:10.1038/329512a0. PMID 2443855. S2CID 19220084.
- Rangarajan, Sneha; Mariuzza, Roy A. (2014-03-17). "T cell receptor bias for MHC: co-evolution or co-receptors?". Cellular and Molecular Life Sciences. 71 (16): 3059–3068. doi:10.1007/s00018-014-1600-9. ISSN 1420-682X. PMID 24633202. S2CID 15214132.
- Yin, Lei; Scott-Browne, James; Kappler, John W.; Gapin, Laurent; Marrack, Philippa (2012-11-01). "T cells and their eons-old obsession with MHC". Immunological Reviews. 250 (1): 49–60. doi:10.1111/imr.12004. ISSN 1600-065X. PMC 3963424. PMID 23046122.
- Flajnik, Martin F.; Kasahara, Masanori (2010). "Origin and evolution of the adaptive immune system: genetic events and selective pressures". Nature Reviews Genetics. 11 (1): 47–59. doi:10.1038/nrg2703. PMC 3805090. PMID 19997068.
- Scott-Browne, James P.; White, Janice; Kappler, John W.; Gapin, Laurent; Marrack, Philippa (2009). "Germline-encoded amino acids in the αβ T-cell receptor control thymic selection". Nature. 458 (7241): 1043–1046. Bibcode:2009Natur.458.1043S. doi:10.1038/nature07812. PMC 2679808. PMID 19262510.
- Deng, Lu; Langley, Ries J.; Wang, Qian; Topalian, Suzanne L.; Mariuzza, Roy A. (2012-09-11). "Structural insights into the editing of germ-line–encoded interactions between T-cell receptor and MHC class II by Vα CDR3". Proceedings of the National Academy of Sciences. 109 (37): 14960–14965. Bibcode:2012PNAS..10914960D. doi:10.1073/pnas.1207186109. ISSN 0027-8424. PMC 3443186. PMID 22930819.
- Van Laethem, François; Tikhonova, Anastasia N.; Pobezinsky, Leonid A.; Tai, Xuguang; Kimura, Motoko Y.; Le Saout, Cécile; Guinter, Terry I.; Adams, Anthony; Sharrow, Susan O. (2013-09-12). "Lck Availability during Thymic Selection Determines the Recognition Specificity of the T Cell Repertoire". Cell. 154 (6): 1326–1341. doi:10.1016/j.cell.2013.08.009. ISSN 0092-8674. PMC 3792650. PMID 24034254.
- Tikhonova, Anastasia N.; Van Laethem, François; Hanada, Ken-ichi; Lu, Jinghua; Pobezinsky, Leonid A.; Hong, Changwan; Guinter, Terry I.; Jeurling, Susanna K.; Bernhardt, Günter (2012-01-27). "αβ T Cell Receptors that Do Not Undergo Major Histocompatibility Complex-Specific Thymic Selection Possess Antibody-like Recognition Specificities". Immunity. 36 (1): 79–91. doi:10.1016/j.immuni.2011.11.013. ISSN 1074-7613. PMC 3268851. PMID 22209676.
- Merwe, P. Anton van der; Cordoba, Shaun-Paul (2011-01-28). "Late Arrival: Recruiting Coreceptors to the T Cell Receptor Complex". Immunity. 34 (1): 1–3. doi:10.1016/j.immuni.2011.01.001. ISSN 1074-7613. PMID 21272780.
External links
 Media related to MHC restriction at Wikimedia Commons
Media related to MHC restriction at Wikimedia Commons