Major royal jelly protein
Major royal jelly proteins (MRJPs) are a family of proteins secreted by honey bees. The family consists of nine proteins, of which MRJP1 (also called royalactin), MRJP2, MRJP3, MRJP4, and MRJP5 are present in the royal jelly secreted by worker bees. MRJP1 is the most abundant, and largest in volume. The five proteins constitute 82–90% of the total proteins in royal jelly.[1][2] Royal jelly is a nutrient-rich mixture of vitamins, sugars, fats, proteins, and enzymes. It is used for feeding larvae.[1] Royal jelly has been used in traditional medicine since ancient times, and the MRJPs are shown to be the main medicinal components. They are synthesised by a family of nine genes (mrjp genes), which are in turn members of the yellow family of genes, such as in the fruitfly (Drosophila) and bacteria. They are involved in the differential development of queen larva and worker larvae, thus establishing division of labour in the bee colony.[1]
| Major royal jelly protein | |||||||
|---|---|---|---|---|---|---|---|
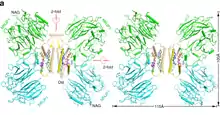 MRJP1 oligomer showing the H-like complex structure that contains four MRJP1 (green or cyan), four apisimin (yellow or wheat), and eight 24-methylenecholesterol (blue, purple, orange, or lime sticks) molecules | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | Mrjp1 | ||||||
| Entrez | 406090 | ||||||
| RefSeq (mRNA) | NM_001011579.1 | ||||||
| RefSeq (Prot) | NP_001011579.1 | ||||||
| UniProt | O18330 | ||||||
| Other data | |||||||
| Chromosome | LG11: 2.57 - 2.57 Mb | ||||||
| |||||||
Discovery
The chemical investigation on royal jelly started in the 1960s.[3] Jozef Hanes and Jozef Šimuth, of the Slovak Academy of Sciences, were the first to identify major royal jelly protein from the hypopharyngeal glands. In 1992 they isolated the protein as a complex of two molecules.[4]
MRJP1 as a single molecule (monomer) was first isolated by Masaki Kamakura and his team at the Toyama Prefectural University in 2001. He found two proteins as potential markers for freshness of royal jelly protein and named them royal jelly proteins (RJP-1 and RJP-2). RJP-1 was a 57-kDa monomer which is a subunit of a larger complex (oligomer).[5] In 2011, Kamakura claimed that RJP-1 is the main protein for controlling larval development that distinguishes the queen from workers. He gave a new name royalactin.[6] This claim has since been challenged.[7]
In 1994, Hanes and Šimuth's team identified genes called pRJP57–1 and pRJP57–2 from the bee head and found that these genes produce similar protein to the first MRJP.[8] By 1999, several independent scientists confirmed the existence of five MRJPs.[3] The Honeybee Genome Sequencing Consortium reported in 2006 that there are nine genes for nine MRJPs.[9]
Structure
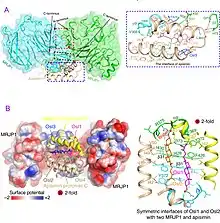
MRJP1 is the most abundant protein in royal jelly. It can exists in two forms, as monomer (single structure) and as oligomer (combined structure). The molecular size of the oligomer is 290-350 kDa. The oligomer is a combination of five monomers.[5] The monomers are associated with another protein apimisin. The monomer is 55 kDa in mass, while apimisin is 5 kDa. The monomer contains 432 amino acids, and is divisible (can be cleaved) into three chains, such as jellein-1, jellein-2, and jellein-4. The monomers in the oligomer are held together by apimisin using noncovalent bonds. The oligomer is resistant to high temperature. MRJP2, MRJP3, MRJP4 and MRJP5 are smaller and their size range between 49 and 80 kDa.[10]
Synthesis
All MRJPs are synthesised from the hypopharyngeal glands, except for MRJP8. MRJP8 is produced in the head of nurse bees, specifically by the Kenyon cells in the mushroom bodies.[11] It was earlier established that MRJP1 to 5 are produced only by young female workers (nurses); hence the genes mrjp1 to 5 are exclusively active in nurses.[12] But later research showed that mrjp genes are expressed also in forager and the queen, not only in their hypopharynx, but also in their brains and abdomen. mrjp1-7 are expressed in the heads of worker bees, with a higher activity of mrjp1-4 and mrjp7 in nurse bees compared to foragers. In contrast, mrjp5 and mrjp6 are more active in foragers compared. mrjp9 is active in the heads, thoraces and abdomen of all female bees. This indicates that mrjp9 is the oldest gene in the family.[13] The mrjp1 gene covers 3038 bp and contains six exons separated by five introns.[14]
Function
As a major component of the royal jelly, MRJPs are the primary proteins in the diet of bee larvae.[15] Other than their nutritional value, their exact biological function is yet to be confirmed.
Controversy
In 2011, Masaki Kamakura published a study claiming that MRJP1 controls division of labour (called polyethism).[6] Kamakura professed to have demonstrated that MRJP1 is the main factor for differentiation of the queen larva from worker larvae. In the queen larva, MRJP1 seemed to induce faster growth, juvenile hormone secretion and development of ovary, while reducing the maturation period.
However, this claim was later challenged by a team of researchers at the Martin Luther University of Halle-Wittenberg in 2016 when they attempted to repeat and expand upon Kamakura's initial study.[7] Despite their larger sample size, they could not replicate Kamakura's findings and called into question the validity of the previous study. They found no difference in queen rearing success between larvae fed normal diets and diets in which the MRJP1 was experimentally degraded and inactivated. This 2016 study reported that MRJP1 alone is not the main protein, but MRJP2, MRJP3, and MRJP5 are equally important in the larval development of the queen, consistent with the theory that queen determination is driven by the total amount of food consumed by a larva.[7]
Additionally the team argued that Kamakura's reported 100% success rate when rearing queens was unrealistic and questionable.[6][7] The Martin Luther University team claimed that it "... is the highest rate ever achieved in over six decades of in vitro queen rearing." The team continued, writing, "Although there is considerable variance in the queen determination rate within and among different laboratories ... it has never, to our knowledge, approached 100%."[7]
Use
MRJPs (as whole royal jelly) are used in the pharmaceutical and cosmetic fields, and are commercialised as an over-the-counter food supplements. They have antimicrobial activities against bacteria, fungi, and viruses. They also show an ability to lower blood pressure, fats in the blood (hypercholesterolemia), stop tumour growth in vitro, and anti-inflammation.[16]
Adverse effect
Royal jelly has been associated with allergic reactions such as contact dermatitis, acute asthma, and anaphylaxis, which can lead to death. In a clinical diagnosis, MRJP1 and MRJP2 are found to be the main allergens.[17] They induce IgE-mediated hypersensitivity reactions thereby causing type 1 hypersensitivity.[18][19]
References
- Buttstedt A, Moritz RF, Erler S (May 2014). "Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family". Biological Reviews of the Cambridge Philosophical Society. 89 (2): 255–69. doi:10.1111/brv.12052. PMID 23855350.
- Albert S, Bhattacharya D, Klaudiny J, Schmitzová J, Simúth J (August 1999). "The family of major royal jelly proteins and its evolution". Journal of Molecular Evolution. 49 (2): 290–7. Bibcode:1999JMolE..49..290A. doi:10.1007/PL00006551. PMID 10441680.
- Buttstedt A, Moritz RF, Erler S (May 2014). "Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family". Biological Reviews of the Cambridge Philosophical Society. 89 (2): 255–69. doi:10.1111/brv.12052. PMID 23855350.
- Hanes J, Šimuth J (2015). "Identification and partial characterization of the major royal jelly protein of the honey bee (Apis mellifera L.)". Journal of Apicultural Research. 31 (1): 22–26. doi:10.1080/00218839.1992.11101256.
- Kamakura M, Fukuda T, Fukushima M, Yonekura M (February 2001). "Storage-dependent degradation of 57-kDa protein in royal jelly: a possible marker for freshness". Bioscience, Biotechnology, and Biochemistry. 65 (2): 277–84. doi:10.1271/bbb.65.277. PMID 11302159.
- Kamakura M (May 2011). "Royalactin induces queen differentiation in honeybees". Nature. 473 (7348): 478–83. Bibcode:2011Natur.473..478K. doi:10.1038/nature10093. hdl:2123/10940. PMID 21516106.
- Buttstedt A, Ihling CH, Pietzsch M, Moritz RF (September 2016). "Royalactin is not a royal making of a queen". Nature. 537 (7621): E10–2. Bibcode:2016Natur.537E..10B. doi:10.1038/nature19349. PMID 27652566.
- Klaudiny J, Hanes J, Kulifajová J, Albert Š, Šimúth J (1994). "Molecular cloning of two cDNAs from the head of the nurse honey bee (Apis mellifera L.) for coding related proteins of royal jelly". Journal of Apicultural Research. 33 (2): 105–111. doi:10.1080/00218839.1994.11100857.
- Honeybee Genome Sequencing Consortium (October 2006). "Insights into social insects from the genome of the honeybee Apis mellifera". Nature. 443 (7114): 931–49. Bibcode:2006Natur.443..931T. doi:10.1038/nature05260. PMC 2048586. PMID 17073008.
- Tamura S, Amano S, Kono T, Kondoh J, Yamaguchi K, Kobayashi S, Ayabe T, Moriyama T (December 2009). "Molecular characteristics and physiological functions of major royal jelly protein 1 oligomer". Proteomics. 9 (24): 5534–43. doi:10.1002/pmic.200900541. PMID 20017154.
- Kucharski R, Maleszka R, Hayward DC, Ball EE (July 1998). "A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain". Die Naturwissenschaften. 85 (7): 343–6. Bibcode:1998NW.....85..343K. doi:10.1007/s001140050512. PMID 9722965.
- Ueno T, Nakaoka T, Takeuchi H, Kubo T (August 2009). "Differential gene expression in the hypopharyngeal glands of worker honeybees (Apis mellifera L.) associated with an age-dependent role change". Zoological Science. 26 (8): 557–63. doi:10.2108/zsj.26.557. PMID 19719408.
- Buttstedt A, Moritz RF, Erler S (November 2013). "More than royal food - Major royal jelly protein genes in sexuals and workers of the honeybee Apis mellifera". Frontiers in Zoology. 10 (1): 72. doi:10.1186/1742-9994-10-72. PMC 4176732. PMID 24279675.
- Malecová B, Ramser J, O'Brien JK, Janitz M, Júdová J, Lehrach H, Simúth J (January 2003). "Honeybee (Apis mellifera L.) mrjp gene family: computational analysis of putative promoters and genomic structure of mrjp1, the gene coding for the most abundant protein of larval food". Gene. 303 (16): 165–75. doi:10.1016/S0378-1119(02)01174-5. PMID 12559578.
- Fujita T, Kozuka-Hata H, Ao-Kondo H, Kunieda T, Oyama M, Kubo T (January 2013). "Proteomic analysis of the royal jelly and characterization of the functions of its derivation glands in the honeybee". Journal of Proteome Research. 12 (1): 404–11. doi:10.1021/pr300700e. PMID 23157659.
- Ramadan MF, Al-Ghamdi A (2012). "Bioactive compounds and health-promoting properties of royal jelly: A review". Journal of Functional Foods. 4 (1): 39–52. doi:10.1016/j.jff.2011.12.007.
- Rosmilah M, Shahnaz M, Patel G, Lock J, Rahman D, Masita A, Noormalin A (December 2008). "Characterization of major allergens of royal jelly Apis mellifera". Tropical Biomedicine. 25 (3): 243–51. PMID 19287364.
- Thien FC, Leung R, Baldo BA, Weiner JA, Plomley R, Czarny D (February 1996). "Asthma and anaphylaxis induced by royal jelly". Clinical and Experimental Allergy. 26 (2): 216–22. doi:10.1111/j.1365-2222.1996.tb00082.x. PMID 8835130.
- Hayashi T, Takamatsu N, Nakashima T, Arita T (2011). "Immunological characterization of honey proteins and identification of MRJP 1 as an IgE-binding protein". Bioscience, Biotechnology, and Biochemistry. 75 (3): 556–60. doi:10.1271/bbb.100778. PMID 21389615.