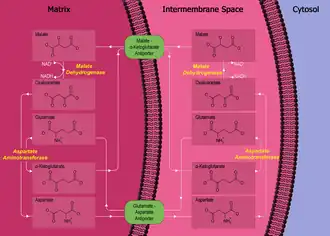
Malate–aspartate shuttle
The malate–aspartate shuttle (sometimes simply the malate shuttle) is a biochemical system for translocating electrons produced during glycolysis across the semipermeable inner membrane of the mitochondrion for oxidative phosphorylation in eukaryotes. These electrons enter the electron transport chain of the mitochondria via reduction equivalents to generate ATP. The shuttle system is required because the mitochondrial inner membrane is impermeable to NADH, the primary reducing equivalent of the electron transport chain. To circumvent this, malate carries the reducing equivalents across the membrane.
Components
The shuttle consists of four protein parts:
- malate dehydrogenase in the mitochondrial matrix and intermembrane space.
- aspartate aminotransferase in the mitochondrial matrix and intermembrane space.
- malate-alpha-ketoglutarate antiporter in the inner membrane.[1]
- glutamate-aspartate antiporter in the inner membrane.[1]
Mechanism
The primary enzyme in the malate–aspartate shuttle is malate dehydrogenase. Malate dehydrogenase is present in two forms in the shuttle system: mitochondrial malate dehydrogenase and cytosolic malate dehydrogenase. The two malate dehydrogenases are differentiated by their location and structure, and catalyze their reactions in opposite directions in this process.
First, in the cytosol, malate dehydrogenase catalyses the reaction of oxaloacetate and NADH to produce malate and NAD+. In this process, two electrons generated from NADH, and an accompanying H+, are attached to oxaloacetate to form malate.
Once malate is formed, the first antiporter (malate-alpha-ketoglutarate) imports the malate from the cytosol into the mitochondrial matrix and also exports alpha-ketoglutarate from the matrix into the cytosol simultaneously. After malate reaches the mitochondrial matrix, it is converted by mitochondrial malate dehydrogenase into oxaloacetate, during which NAD+ is reduced with two electrons to form NADH. Oxaloacetate is then transformed into aspartate (since oxaloacetate cannot be transported into the cytosol) by mitochondrial aspartate aminotransferase. Since aspartate is an amino acid, an amino radical needs to be added to the oxaloacetate. This is supplied by glutamate, which in the process is transformed into alpha-ketoglutarate by the same enzyme.
The second antiporter (the glutamate-aspartate antiporter) imports glutamate from the cytosol into the matrix and exports aspartate from the matrix to the cytosol. Once in the cytosol, aspartate is converted by cytosolic aspartate aminotransferase to oxaloacetate.
The net effect of the malate–aspartate shuttle is purely redox: NADH in the cytosol is oxidized to NAD+, and NAD+ in the matrix is reduced to NADH. The NAD+ in the cytosol can then be reduced again by another round of glycolysis, and the NADH in the matrix can be used to pass electrons to the electron transport chain so ATP can be synthesized.
Since the malate–aspartate shuttle regenerates NADH inside the mitochondrial matrix, it is capable of maximizing the number of ATPs produced in glycolysis (3/NADH), ultimately resulting in a net gain of 38 ATP molecules per molecule of glucose metabolized. Compare this to the glycerol 3-phosphate shuttle, which reduces FAD+ to produce FADH2, donates electrons to the quinone pool in the electron transport chain, and is capable of generating only 2 ATPs per NADH generated in glycolysis (ultimately resulting in a net gain of 36 ATPs per glucose metabolized). (These ATP numbers are prechemiosmotic, and should be reduced in light of the work of Mitchell and many others. Each NADH produces only 2.5 ATPs, and each FADH2 produces only 1.5 ATPs. Hence, the ATPs per glucose should be reduced to 32 from 38 and 30 from 36. The extra H+ required to bring in the inorganic phosphate during oxidative-phosphorylation contributes to the 30 and 32 numbers as well).
Regulation
The activity of malate–aspartate shuttle is modulated by arginine methylation of malate dehydrogenase 1 (MDH1). Protein arginine N-methyltransferase CARM1 methylates and inhibits MDH1 by disrupting its dimerization, which represses malate–aspartate shuttle and inhibits mitochondria respiration of pancreatic cancer cells.[2]
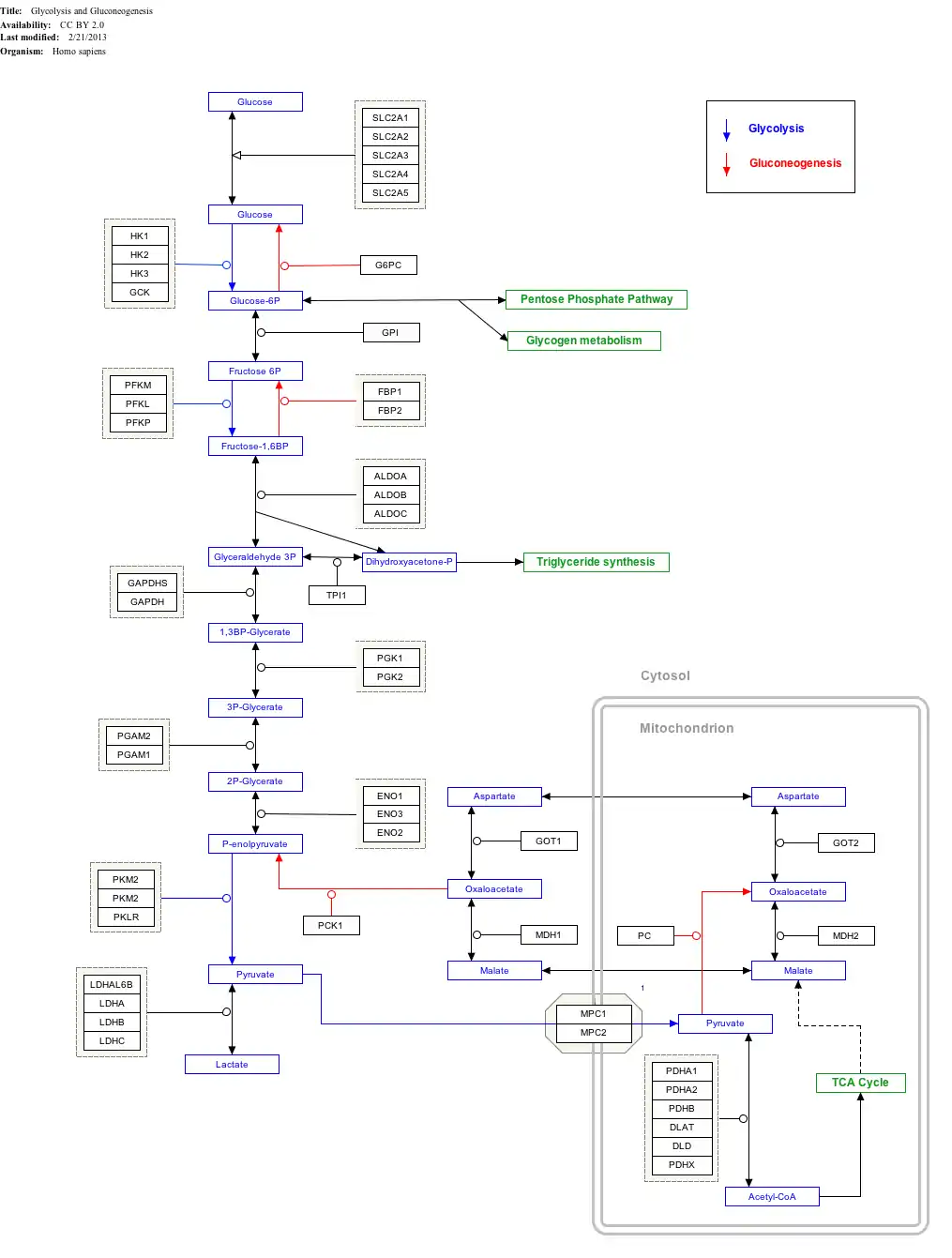
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
References
- Lu, M; Zhou, L; Stanley, WC; Cabrera, ME; Saidel, GM; Yu, X (2008). "Role of the malate–aspartate shuttle on the metabolic response to myocardial ischemia". J. Theor. Biol. 254 (2): 466–75. Bibcode:2008JThBi.254..466L. doi:10.1016/j.jtbi.2008.05.033. PMC 2572303. PMID 18603266.
- Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS, Gao X, Xu YY, Zou SW, Liu YB, Cheng JK, Lei QY (Nov 2016). "Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer". Molecular Cell. 64 (4): 673–87. doi:10.1016/j.molcel.2016.09.028. PMID 27840030.
- Monty Krieger; Matthew P Scott; Matsudaira, Paul T.; Lodish, Harvey F.; Darnell, James E.; Lawrence Zipursky; Kaiser, Chris; Arnold Berk (2003). Molecular Cell Biology, Fifth Edition. San Francisco: W. H. Freeman. ISBN 0-7167-4366-3.