Methyl-CpG-binding domain
The Methyl-CpG-binding domain (MBD) in molecular biology binds to DNA that contains one or more symmetrically methylated CpGs.[1] MBD has negligible non-specific affinity for unmethylated DNA. In vitro foot-printing with the chromosomal protein MeCP2 showed that the MBD could protect a 12 nucleotide region surrounding a methyl CpG pair.[1]
| MBD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
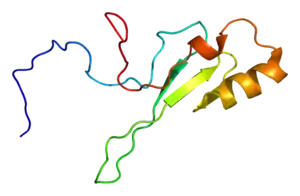 solution structure of human MECO2 | |||||||||
| Identifiers | |||||||||
| Symbol | MBD | ||||||||
| Pfam | PF01429 | ||||||||
| Pfam clan | CL0081 | ||||||||
| InterPro | IPR001739 | ||||||||
| SCOP2 | 1qk9 / SCOPe / SUPFAM | ||||||||
| CDD | cd00122 | ||||||||
| |||||||||
DNA methylation at CpG dinucleotides, the most common DNA modification in eukaryotes, has been associated with various phenomena such as alterations in chromatin structure, genomic imprinting, transposon and chromosome X inactivation, differentiation, and cancer. Effects of DNA methylation are mediated through proteins that bind to symmetrically methylated CpGs. Such proteins contain a specific domain of ~70 residues, the methyl-CpG-binding domain (MBD), which is linked to additional domains associated with chromatin, such as the bromodomain, the AT hook motif, the SET domain, or the PHD finger. MBD-containing proteins appear to act as structural proteins, which recruit a variety of histone deacetylase (HDAC) complexes and chromatin remodelling factors, leading to chromatin compaction and, consequently, to transcriptional repression. The MBD of MeCP2, MBD1, MBD2, MBD4 and BAZ2 mediates binding to DNA, and in cases of MeCP2, MBD1 and MBD2, preferentially to methylated CpG. In human MBD3 and SETDB1, the MBD has been shown to mediate protein-protein interactions.[2][3] MBDs are also found in DNA demethylase.[4]
The MBD folds into an alpha/beta sandwich structure comprising a layer of twisted beta sheet, backed by another layer formed by the alpha1 helix and a hairpin loop at the C terminus. These layers are both amphipathic, with the alpha1 helix and the beta sheet lying parallel and the hydrophobic faces tightly packed against each other. The beta sheet is composed of two long inner strands (beta2 and beta3) sandwiched by two shorter outer strands (beta1 and beta4).[5]
The structure of the MBD domain bound to methylated DNA has been solved (PDB: 3C2I). It recognizes the hydration of the major groove at methylated sites.[6]
References
- Nan X, Meehan RR, Bird A (1993). "Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2". Nucleic Acids Res. 21 (21): 4886–92. doi:10.1093/nar/21.21.4886. PMC 311401. PMID 8177735.
- Roloff TC, Ropers HH, Nuber UA (January 2003). "Comparative study of methyl-CpG-binding domain proteins". BMC Genomics. 4 (1): 1. doi:10.1186/1471-2164-4-1. PMC 149351. PMID 12529184.
- Zemach A, Grafi G (June 2003). "Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins". Plant J. 34 (5): 565–72. doi:10.1046/j.1365-313x.2003.01756.x. PMID 12787239.
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M (1999). "A mammalian protein with specific demethylase activity for mCpG DNA". Nature. 397 (6720): 579–83. Bibcode:1999Natur.397..579B. doi:10.1038/17533. PMID 10050851. S2CID 4408031.
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M (May 2001). "Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA". Cell. 105 (4): 487–97. doi:10.1016/S0092-8674(01)00324-5. PMID 11371345.
- Ho, KL; McNae, IW; Schmiedeberg, L; Klose, RJ; Bird, AP; Walkinshaw, MD (29 February 2008). "MeCP2 binding to DNA depends upon hydration at methyl-CpG". Molecular Cell. 29 (4): 525–31. doi:10.1016/j.molcel.2007.12.028. PMID 18313390.