Intelectin
Intelectins are lectins (carbohydrate-binding proteins) expressed in humans and other chordates. Humans express two types of intelectins encoded by ITLN1 and ITLN2 genes respectively.[1][2] Several intelectins bind microbe-specific carbohydrate residues. Therefore, intelectins have been proposed to function as immune lectins.[3][4] Even though intelectins contain fibrinogen-like domain found in the ficolins family of immune lectins, there is significant structural divergence.[5] Thus, intelectins may not function through the same lectin-complement pathway. Most intelectins are still poorly characterized and they may have diverse biological roles. Human intelectin-1 (hIntL-1) has also been shown to bind lactoferrin,[6] but the functional consequence has yet to be elucidated. Additionally, hIntL-1 is a major component of asthmatic mucus[7] and may be involved in insulin physiology as well.[8]
| Xenopus embryonic epidermal lectin | |||||||
|---|---|---|---|---|---|---|---|
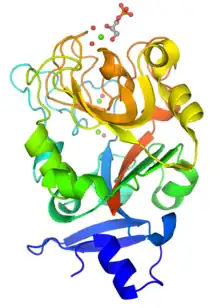 Monomeric structure of XEEL-CRD with bound D-glycerol 1-phosphate. The protein is colored using a blue-red gradient from the N- to the C- terminus. Calcium ions are shown as green spheres and the coordinated water molecules are shown as red spheres. | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | itln1 | ||||||
| Entrez | 398574 | ||||||
| HomoloGene | 111044 | ||||||
| PDB | 4WN0 | ||||||
| RefSeq (mRNA) | NM_001089101.1 | ||||||
| RefSeq (Prot) | NP_001082570.1 | ||||||
| UniProt | Q800K0 | ||||||
| |||||||
| Human intelectin-1 | |||||||
|---|---|---|---|---|---|---|---|
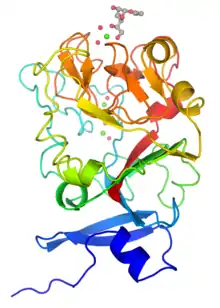 Monomeric structure of human intelectin with bound allyl-beta-D-galactofuranose. The protein is colored using a blue-red gradient from the N- to the C- terminus. Calcium ions are shown as green spheres and the coordinated water molecules are shown as red spheres. | |||||||
| Identifiers | |||||||
| Symbol | ITLN1 | ||||||
| Alt. symbols | hIntL-1 | ||||||
| NCBI gene | 55600 | ||||||
| HGNC | 18259 | ||||||
| OMIM | 609873 | ||||||
| PDB | 4WMY | ||||||
| RefSeq | NP_060095 | ||||||
| UniProt | Q8WWA0 | ||||||
| Other data | |||||||
| Locus | Chr. 1 q21.3 | ||||||
| |||||||
Diversity
The first intelectin was discovered in Xenopus laevis oocyte and is named XL35 or XCGL-1.[9][10][11] X. laevis oocyte also contains a closely related XCGL-2.[12] In addition, X. laevis embryos secrete Xenopus embryonic epidermal lectin into the environmental water, presumably to bind microbes.[13][14] XSL-1 and XSL-2 are also expressed in X. laevis serum when stimulated with lipopolysaccharide.[15] Two additional intestinal intelectins are discovered in X. laevis[16]
Human has two intelectins: hIntL-1 (omentin) and hIntL-2.[17] Mouse also has two intelectins: mIntL-1 and mIntL-2.[18]
Immune system
Several lines of evidence suggest that intelectins recognize microbes and may function as an innate immune defense protein. Tunicate intelectin is an opsonin for phagocytosis by hemocyte.[19] Amphioxus intelectin has been shown to agglutinate bacteria.[20][21] In zebrafish and rainbow trout, intelectin expression is stimulated upon microbial exposure.[22][23][24] Mammals such as sheep and mice also upregulate intelectin expression upon parasitic infection.[25][26] Increase in intelectin expression upon microbial exposure support the hypothesis that intelectins play a role in the immune system.
Structure
Although intelectins require calcium ion for function, the sequences bear no resemblance to C-type lectins.[3] In addition, merely around 50 amino acids (the fibronogen-like domain) align with any known protein, specifically the ficolin family.[2] The first structural details of an intelectin comes from the crystal structure of selenomethionine-labeled XEEL carbohydrate-recognition domain (Se-Met XEEL-CRD) solved by Se-SAD.[5] XEEL-CRD was expressed and Se-Met-labeled in High Five insect cells using a recombinant baculovirus. The fibrinogen-like fold is conserved despite amino acid sequence divergence. However, extensive insertions are present in intelectin compared to ficolins, thus making intelectin a distinct lectin structural class.[5] The Se-Met XEEL-CRD structure then enables the structure solution by molecular replacement of D-glycerol 1-phosphate (GroP)-bound XEEL-CRD,[5] apo-human intelectin-1 (hIntL-1),[4] and galactofuranose-bound hIntL-1.[4]
Each polypeptide chain of XEEL and hIntL-1 contains three bound calcium ions: two in the structural calcium site and one in the ligand binding site.[4][5] The amino acid residues in the structural calcium site are conserved among intelectins, thus it is likely that most, if not all, intelectins have two structural calcium ions.[5]
In the ligand binding site of XEEL and hIntL-1, the exocyclic vicinal diol of the carbohydrate ligand directly coordinates to the calcium ion.[4][5] There are large variations in the ligand binding site residues among intelectin homologs suggesting that the intelectin family may have broad ligand specificities and biological functions.[5] As there is no intelectin numbering conventions in different organisms, one should not assume functional homology based on the intelectin number. For example, hIntL-1 has glutamic acid residues in the ligand binding site to coordinate a calcium ion, while zebrafish intelectin-1 are devoided of these acidic residues.[5] Zebrafish intelectin-2 ligand binding site residues are similar to those present in hIntL-1.
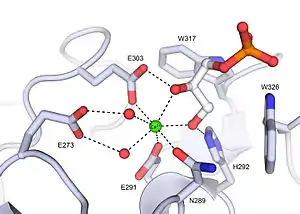 Xenopus embryonic epidermal lectin (XEEL) ligand binding site with bound D-glycerol 1-phosphate. The calcium ion is shown as a green sphere and the ordered water molecules are shown as red spheres.[5]
Xenopus embryonic epidermal lectin (XEEL) ligand binding site with bound D-glycerol 1-phosphate. The calcium ion is shown as a green sphere and the ordered water molecules are shown as red spheres.[5]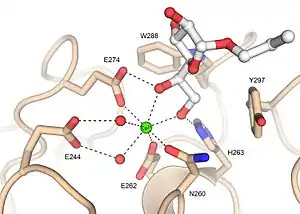 Human intelectin-1 (hIntL-1) ligand binding site with bound allyl-beta-D-galactofuranose. The calcium ion is shown as a green sphere and the ordered water molecules are shown as red spheres.[4]
Human intelectin-1 (hIntL-1) ligand binding site with bound allyl-beta-D-galactofuranose. The calcium ion is shown as a green sphere and the ordered water molecules are shown as red spheres.[4]
Oligomeric state
hIntL-1 is a disulfide-linked trimer as shown by non-reducing SDS-PAGE[3] and X-ray crystallography.[4] Despite lacking the intermolecular disulfide bonds, XEEL-CRD is trimeric in solution.[5] The N-terminal peptide of the full length XEEL is responsible for dimerizing the trimeric XEEL-CRD into a disulfide-linked hexameric full-length XEEL.[5] Therefore, the N-termini of intelectins are often responsible for forming disulfide-linked oligomer. In intelectin homologs where the N-terminal cysteines are absent, the CRD itself may still capable of forming non-covalent oligomer in solution.
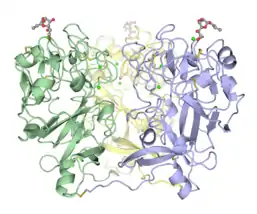 Disulfide-linked trimeric human intelectin-1.[4]
Disulfide-linked trimeric human intelectin-1.[4]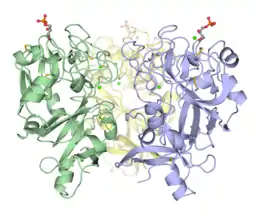 Trimeric Xenopus embryonic epidermal lectin carbohydrate-recognition domain (XEEL-CRD). Extensive biophysical investigations conclusively indicate that XEEL-CRD is trimeric in solution despite lacking the intermolecular disulfide bonds found in hIntL-1.[5]
Trimeric Xenopus embryonic epidermal lectin carbohydrate-recognition domain (XEEL-CRD). Extensive biophysical investigations conclusively indicate that XEEL-CRD is trimeric in solution despite lacking the intermolecular disulfide bonds found in hIntL-1.[5]
References
- Lee JK, Baum LG, Moremen K, Pierce M (August 2004). "The X-lectins: a new family with homology to the Xenopus laevis oocyte lectin XL-35". Glycoconjugate Journal. 21 (8–9): 443–50. CiteSeerX 10.1.1.537.3931. doi:10.1007/s10719-004-5534-6. PMID 15750785. S2CID 41789407.
- Yan J, Xu L, Zhang Y, Zhang C, Zhang C, Zhao F, Feng L (Oct 2013). "Comparative genomic and phylogenetic analyses of the intelectin gene family: implications for their origin and evolution". Developmental and Comparative Immunology. 41 (2): 189–99. doi:10.1016/j.dci.2013.04.016. PMID 23643964.
- Tsuji S, Uehori J, Matsumoto M, Suzuki Y, Matsuhisa A, Toyoshima K, Seya T (Jun 2001). "Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall". The Journal of Biological Chemistry. 276 (26): 23456–63. doi:10.1074/jbc.M103162200. PMID 11313366.
- Wesener DA, Wangkanont K, McBride R, Song X, Kraft MB, Hodges HL, Zarling LC, Splain RA, Smith DF, Cummings RD, Paulson JC, Forest KT, Kiessling LL (Aug 2015). "Recognition of microbial glycans by human intelectin-1". Nature Structural & Molecular Biology. 22 (8): 603–10. doi:10.1038/nsmb.3053. PMC 4526365. PMID 26148048.
- Wangkanont K, Wesener DA, Vidani JA, Kiessling LL, Forest KT (Jan 2016). "Structures of Xenopus embryonic epidermal lectin reveal a conserved mechanism of microbial glycan recognition". The Journal of Biological Chemistry. 291 (11): 5596–610. doi:10.1074/jbc.M115.709212. PMC 4786701. PMID 26755729.
- Suzuki YA, Shin K, Lönnerdal B (Dec 2001). "Molecular cloning and functional expression of a human intestinal lactoferrin receptor". Biochemistry. 40 (51): 15771–9. doi:10.1021/bi0155899. PMID 11747454.
- Kerr SC, Carrington SD, Oscarson S, Gallagher ME, Solon M, Yuan S, Ahn JN, Dougherty RH, Finkbeiner WE, Peters MC, Fahy JV (Apr 2014). "Intelectin-1 is a prominent protein constituent of pathologic mucus associated with eosinophilic airway inflammation in asthma". American Journal of Respiratory and Critical Care Medicine. 189 (8): 1005–7. doi:10.1164/rccm.201312-2220LE. PMC 4098098. PMID 24735037.
- Yang RZ, Lee MJ, Hu H, Pray J, Wu HB, Hansen BC, Shuldiner AR, Fried SK, McLenithan JC, Gong DW (Jun 2006). "Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action". American Journal of Physiology. Endocrinology and Metabolism. 290 (6): E1253–61. doi:10.1152/ajpendo.00572.2004. PMID 16531507.
- Roberson MM, Barondes SH (Jul 1982). "Lectin from embryos and oocytes of Xenopus laevis. Purification and properties". The Journal of Biological Chemistry. 257 (13): 7520–4. doi:10.1016/S0021-9258(18)34409-0. PMID 7085636.
- Nishihara T, Wyrick RE, Working PK, Chen YH, Hedrick JL (Oct 1986). "Isolation and characterization of a lectin from the cortical granules of Xenopus laevis eggs". Biochemistry. 25 (20): 6013–20. doi:10.1021/bi00368a027. PMID 3098282.
- Lee JK, Buckhaults P, Wilkes C, Teilhet M, King ML, Moremen KW, Pierce M (Apr 1997). "Cloning and expression of a Xenopus laevis oocyte lectin and characterization of its mRNA levels during early development". Glycobiology. 7 (3): 367–72. doi:10.1093/glycob/7.3.367. PMID 9147045.
- Shoji H, Ikenaka K, Nakakita S, Hayama K, Hirabayashi J, Arata Y, Kasai K, Nishi N, Nakamura T (Jul 2005). "Xenopus galectin-VIIa binds N-glycans of members of the cortical granule lectin family (xCGL and xCGL2)". Glycobiology. 15 (7): 709–20. doi:10.1093/glycob/cwi051. PMID 15761024.
- Nagata S, Nakanishi M, Nanba R, Fujita N (Jul 2003). "Developmental expression of XEEL, a novel molecule of the Xenopus oocyte cortical granule lectin family". Development Genes and Evolution. 213 (7): 368–70. doi:10.1007/s00427-003-0341-9. PMID 12802587. S2CID 41996445.
- Nagata S (Mar 2005). "Isolation, characterization, and extra-embryonic secretion of the Xenopus laevis embryonic epidermal lectin, XEEL". Glycobiology. 15 (3): 281–90. doi:10.1093/glycob/cwi010. PMID 15537792.
- Nagata S, Nishiyama S, Ikazaki Y (Jun 2013). "Bacterial lipopolysaccharides stimulate production of XCL1, a calcium-dependent lipopolysaccharide-binding serum lectin, in Xenopus laevis". Developmental and Comparative Immunology. 40 (2): 94–102. doi:10.1016/j.dci.2013.02.008. PMID 23454582.
- Nagata S (Feb 2016). "Identification and characterization of a novel intelectin in the digestive tract of Xenopus laevis". Developmental and Comparative Immunology. 59: 229–239. doi:10.1016/j.dci.2016.02.006. PMID 26855011.
- Lee JK, Schnee J, Pang M, Wolfert M, Baum LG, Moremen KW, Pierce M (Jan 2001). "Human homologs of the Xenopus oocyte cortical granule lectin XL35". Glycobiology. 11 (1): 65–73. doi:10.1093/glycob/11.1.65. PMID 11181563.
- Lu ZH, di Domenico A, Wright SH, Knight PA, Whitelaw CB, Pemberton AD (2011). "Strain-specific copy number variation in the intelectin locus on the 129 mouse chromosome 1". BMC Genomics. 12 (1): 110. doi:10.1186/1471-2164-12-110. PMC 3048546. PMID 21324158.
- Abe Y, Tokuda M, Ishimoto R, Azumi K, Yokosawa H (Apr 1999). "A unique primary structure, cDNA cloning and function of a galactose-specific lectin from ascidian plasma". European Journal of Biochemistry. 261 (1): 33–9. doi:10.1046/j.1432-1327.1999.00238.x. PMID 10103030.
- Yan J, Wang J, Zhao Y, Zhang J, Bai C, Zhang C, Zhang C, Li K, Zhang H, Du X, Feng L (Jul 2012). "Identification of an amphioxus intelectin homolog that preferably agglutinates gram-positive over gram-negative bacteria likely due to different binding capacity to LPS and PGN". Fish & Shellfish Immunology. 33 (1): 11–20. doi:10.1016/j.fsi.2012.03.023. PMID 22475783. S2CID 35820556.
- Yan J, Zhang C, Zhang Y, Li K, Xu L, Guo L, Kong Y, Feng L (May 2013). "Characterization and comparative analyses of two amphioxus intelectins involved in the innate immune response". Fish & Shellfish Immunology. 34 (5): 1139–46. doi:10.1016/j.fsi.2013.01.017. PMID 23428515.
- Lin B, Cao Z, Su P, Zhang H, Li M, Lin Y, Zhao D, Shen Y, Jing C, Chen S, Xu A (Mar 2009). "Characterization and comparative analyses of zebrafish intelectins: highly conserved sequences, diversified structures and functions". Fish & Shellfish Immunology. 26 (3): 396–405. doi:10.1016/j.fsi.2008.11.019. PMID 19100836.
- Russell S, Young KM, Smith M, Hayes MA, Lumsden JS (Jul 2008). "Identification, cloning and tissue localization of a rainbow trout (Oncorhynchus mykiss) intelectin-like protein that binds bacteria and chitin". Fish & Shellfish Immunology. 25 (1–2): 91–105. doi:10.1016/j.fsi.2008.02.018. PMID 18502147.
- Russell S, Hayes MA, Lumsden JS (Jan 2009). "Immunohistochemical localization of rainbow trout ladderlectin and intelectin in healthy and infected rainbow trout (Oncorhynchus mykiss)". Fish & Shellfish Immunology. 26 (1): 154–63. doi:10.1016/j.fsi.2008.03.001. PMID 19046637.
- Datta R, deSchoolmeester ML, Hedeler C, Paton NW, Brass AM, Else KJ (Jul 2005). "Identification of novel genes in intestinal tissue that are regulated after infection with an intestinal nematode parasite". Infection and Immunity. 73 (7): 4025–33. doi:10.1128/IAI.73.7.4025-4033.2005. PMC 1168561. PMID 15972490.
- French AT, Knight PA, Smith WD, Brown JK, Craig NM, Pate JA, Miller HR, Pemberton AD (Mar 2008). "Up-regulation of intelectin in sheep after infection with Teladorsagia circumcincta". International Journal for Parasitology. 38 (3–4): 467–75. doi:10.1016/j.ijpara.2007.08.015. PMID 17983620.
Further reading
- Wesener DA, Wangkanont K, McBride R, Song X, Kraft MB, Hodges HL, Zarling LC, Splain RA, Smith DF, Cummings RD, Paulson JC, Forest KT, Kiessling LL (Aug 2015). "Recognition of microbial glycans by human intelectin-1". Nature Structural & Molecular Biology. 22 (8): 603–10. doi:10.1038/nsmb.3053. PMC 4526365. PMID 26148048. for exhaustive ligand binding analysis of human intelectin-1 (hIntL-1). The article also reveals how hIntL-1 could discriminate between microbial and mammalian cells.
- Wangkanont K, Wesener DA, Vidani JA, Kiessling LL, Forest KT (Jan 2016). "Structures of Xenopus embryonic epidermal lectin reveal a conserved mechanism of microbial glycan recognition". The Journal of Biological Chemistry. 291 (11): 5596–610. doi:10.1074/jbc.M115.709212. PMC 4786701. PMID 26755729. for discussion on how the first intelectin structure (XEEL-CRD) was solved. In depth biophysical and evolutionary analyses of the intelectin family in the light of the available 3D structures also provide significant insights into this protein family not previously appreciated. The article serves as the most up-to-date review on the biochemistry of the intelectin family.
- Yan J, Xu L, Zhang Y, Zhang C, Zhang C, Zhao F, Feng L (Oct 2013). "Comparative genomic and phylogenetic analyses of the intelectin gene family: implications for their origin and evolution". Developmental and Comparative Immunology. 41 (2): 189–99. doi:10.1016/j.dci.2013.04.016. PMID 23643964. for comprehensive genomics analysis of intelectins from various organisms.