Bacterial outer membrane
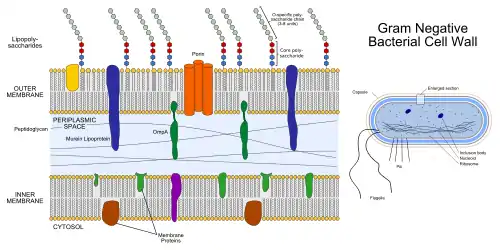
The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.
| Lipopolysaccharide-assembly, LptC-related | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | LptC | ||||||||
| Pfam | PF06835 | ||||||||
| Pfam clan | CL0259 | ||||||||
| InterPro | IPR010664 | ||||||||
| |||||||||
| Lipopolysaccharide-assembly | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | LptE | ||||||||
| Pfam | PF04390 | ||||||||
| InterPro | IPR007485 | ||||||||
| TCDB | 1.B.42 | ||||||||
| OPM superfamily | 412 | ||||||||
| OPM protein | 4q35 | ||||||||
| |||||||||
Clinical significance
If lipid A, part of the lipopolysaccharide, enters the circulatory system it causes a toxic reaction by activating toll like receptor TLR 4. Lipid A is very pathogenic and not immunogenic. However, the polysaccharide component is very immunogenic, but not pathogenic, causing an aggressive response by the immune system. The sufferer will have a high temperature and respiration rate and a low blood pressure. This may lead to endotoxic shock, which may be fatal. The bacterial outer membrane is physiologically shed as the bounding membrane of outer membrane vesicles in cultures, as well as in animal tissues at the host-pathogen interface, implicated in translocation of gram-negative microbial biochemical signals to host or target cells.
Biogenesis
The biogenesis of the outer membrane requires that the individual components are transported from the site of synthesis to their final destination outside the inner membrane by crossing both hydrophilic and hydrophobic compartments. The machinery and the energy source that drive this process are not yet fully understood. The lipid A-core moiety and the O-antigen repeat units are synthesized at the cytoplasmic face of the inner membrane and are separately exported via two independent transport systems, namely, the O-antigen transporter Wzx (RfbX) and the ATP binding cassette (ABC) transporter MsbA that flips the lipid A-core moiety from the inner leaflet to the outer leaflet of the inner membrane.[2][3][4][5][6] O-antigen repeat units are then polymerised in the periplasm by the Wzy polymerase and ligated to the lipid A-core moiety by the WaaL ligase.[7][8]
The LPS transport machinery is composed of LptA, LptB, LptC, LptD, LptE. This supported by the fact that depletion of any one of these proteins blocks the LPS assembly pathway and results in very similar outer membrane biogenesis defects. Moreover, the location of at least one of these five proteins in every cellular compartment suggests a model for how the LPS assembly pathway is organised and ordered in space.[8]
LptC is required for the translocation of lipopolysaccharide (LPS) from the inner membrane to the outer membrane.[8] LptE forms a complex with LptD, which is involved in the assembly of LPS in the outer leaflet of the outer membrane and is essential for envelope biogenesis.[8][9][10]
References
- van der Ley P, Heckels JE, Virji M, Hoogerhout P, Poolman JT (September 1991). "Topology of outer membrane porins in pathogenic Neisseria spp". Infection and Immunity. 59 (9): 2963–71. doi:10.1128/IAI.59.9.2963-2971.1991. PMC 258120. PMID 1652557.
- Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA (December 1999). "The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat". J. Biol. Chem. 274 (49): 35129–38. doi:10.1074/jbc.274.49.35129. PMID 10574995.
- Liu D, Cole RA, Reeves PR (April 1996). "An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase". J. Bacteriol. 178 (7): 2102–7. doi:10.1128/jb.178.7.2102-2107.1996. PMC 177911. PMID 8606190.
- Doerrler WT, Reedy MC, Raetz CR (April 2001). "An Escherichia coli mutant defective in lipid export". J. Biol. Chem. 276 (15): 11461–4. doi:10.1074/jbc.C100091200. PMID 11278265.
- Polissi A, Georgopoulos C (June 1996). "Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter". Mol. Microbiol. 20 (6): 1221–33. doi:10.1111/j.1365-2958.1996.tb02642.x. PMID 8809774. S2CID 26807796.
- Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR (May 1998). "Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis". J. Biol. Chem. 273 (20): 12466–75. doi:10.1074/jbc.273.20.12466. hdl:2434/611267. PMID 9575204.
- Raetz CR, Whitfield C (2002). "Lipopolysaccharide endotoxins". Annu. Rev. Biochem. 71: 635–700. doi:10.1146/annurev.biochem.71.110601.135414. PMC 2569852. PMID 12045108.
- Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A (July 2008). "Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli". J. Bacteriol. 190 (13): 4460–9. doi:10.1128/JB.00270-08. PMC 2446812. PMID 18424520.
- Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D (August 2006). "Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli". Proc. Natl. Acad. Sci. U.S.A. 103 (31): 11754–9. Bibcode:2006PNAS..10311754W. doi:10.1073/pnas.0604744103. PMC 1544242. PMID 16861298.
- Bos MP, Tefsen B, Geurtsen J, Tommassen J (June 2004). "Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface". Proc. Natl. Acad. Sci. U.S.A. 101 (25): 9417–22. Bibcode:2004PNAS..101.9417B. doi:10.1073/pnas.0402340101. PMC 438991. PMID 15192148.