Oxidation response
Oxidation response is stimulated by a disturbance in the balance between the production of reactive oxygen species and antioxidant responses, known as oxidative stress. Active species of oxygen naturally occur in aerobic cells and have both intracellular and extracellular sources. These species, if not controlled, damage all components of the cell, including proteins, lipids and DNA. Hence cells need to maintain a strong defense against the damage. The following table gives an idea of the antioxidant defense system in bacterial system.
| Line of defense | Components | Function | Examples |
|---|---|---|---|
First | Metal chelators | prevent free radical formation by inhibiting metal catalyzed reactions | |
Second | Low molecular weight compounds and antioxidant enzymes | deactivate free radicals (ROS) before some biological molecule is damaged |
superoxide dismutase (SOD) and catalase |
Third |
DNA repair systems
protein repair system lipid repair system |
repair biomolecules after they were damaged by ROS |
Stress response
Small changes in cellular oxidant status can be sensed by specific proteins which regulate a set of genes encoding antioxidant enzymes. Such a global response induces an adaptive metabolism including ROS elimination, the bypass of injured pathways, reparation of oxidative damages and maintenance of reducing power.
Peroxide and superoxide are the two major active oxygen species. It is found that the peroxide and superoxide stress responses are distinct in bacteria. The exposure of microorganisms to low sublethal concentrations of oxidants leads to the acquisition of cellular resistance to a subsequent lethal oxidative stress.
Peroxide stress response
In response to an increased flux of hydrogen peroxide and other organic peroxides such as tert-butyl hydroperoxide and cumene hydroperoxide, peroxide stimulon gets activated. Studies of E. coli response to H2O2 have shown that exposure to H2O2 elevated mRNA levels of 140 genes, of which 30 genes are members of the OxyR regulon. The genes include many genes coding for metabolic enzymes and antioxidant enzymes demonstrating the role of these enzymes in reorganization of metabolism under stress conditions.[1]
Superoxide stress response
When stressed under elevated levels of the superoxide radical anion O2−, bacteria respond by invoking the superoxide stimulon. Superoxide-generating compounds activate SoxR regulator by the one-electron oxidation of the 2Fe-2S clusters. Oxidized SoxR then induces the expression of SoxS protein, which in turn activates the transcription of structural genes of the SoxRS regulon.[2]
Regulation
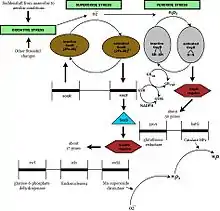
The transcriptional factor OxyR regulates the expression of OxyR regulon. H2O2 oxidizes the transcriptional factor by forming an intramolecular disulfide bond. The oxidized form of this factor specifically binds to the promoters of constituent genes of OxyR regulon, including katG (hydroperoxidase-catalase HPІ), gorA (glutathione reductase), grxA (glutaredoxin 1), trxC(thioredoxin 2), ahpCF (alkyl hydroperoxide reductase), dps (nonspecific DNA binding protein) and oxyS (a small regulatory RNA). Reduced OxyR provides autorepression by binding only to the oxyR promoter.[1]
Regulation of the soxRS regulon occurs by a two-stage process: the SoxR protein is first converted to an oxidized form that enhances soxS transcription, and the increased level of SoxS protein in turn activates the expression of the regulon. The structural genes under this regulon include sodA (Mn-superoxide dismutase(SOD)), zwf (glucose-6-phosphate dehydrogenase(G6PDH)), acnA (aconitase A), nfsA (nitrate reductase A), fumC (fumarase C) and nfo (endonuclease IV) among others. In E.coli, negative autoregulation of SoxS protein serves as a dampening mechanism for the soxRS redox stress response.[3]
SoxRS regulon genes can be regulated by additional factors.[2]
At least three known genes including xthA and katE are regulated by a sigma factor, KatF(RpoS), whose synthesis is turned on during the stationary phase. XthA (exonuclease III, a DNA repair enzyme) and KatE (catalase) are known to play important roles in the defense against oxidative stress but KatF regulon genes are not induced by oxidative stress.[2]
There is an overlap between oxidative stress response and other regulatory networks like heat shock response, SOS response.
Physiological role of the response
The defenses against deleterious effects of active oxygen can be logically divided into two broad classes, preventive and reparative.
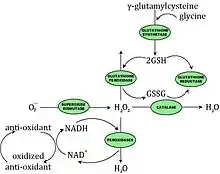
Prevention of Oxidative Damage
Cellular defenses against the damaging effects of oxidative stress involve both enzymatic and nonenzymatic components.
The enzymatic components may directly scavenge active oxygen species or may act by producing the nonenzymatic antioxidants. There are four enzymes that provide the bulk of protection against deleterious reactions involving active oxygen in bacteria: SODs (superoxide dismutases encoded by sodA and sodB), catalases (katE and katG), glutathione synthetase (gshAB) and glutathione reductase (gor). Some bacteria have NADH-dependent peroxidases specific for H2O2.
The main nonenzymatic antioxidants in E. coli are GSH and thioredoxin (encoded by trxA). Ubiquinone and menaquinone may also serve as membrane-associated antioxidants.
Repair of Oxidative damage
Secondary defenses include DNA-repair systems, proteolytic and lipolytic enzymes. DNA repair enzymes include endonuclease IV, induced by oxidative stress, and exonuclease III, induced in the stationary phase and in starving cells. These enzymes act on duplex DNA and clean up DNA 3' terminal ends.
Prokaryotic cells contain catalysts that modify the primary structure of proteins frequently by reducing disulfide bonds. This occurs in the following steps:
(i) thioredoxin reductase transfers electrons from NADPH to thioredoxin via a flavin carrier
(ii) glutaredoxin is also able to reduce disulfide bonds, but using GSH as an electron donor
(iii) protein disulfide isomerase facilitates disulfide exchange reactions with large inactive protein substrates, besides having chaperone activity
Oxidation of surface exposed methionine residues surrounding the entrance to the active site could function as a “last-chance” antioxidant defense system for proteins.[4]
Eukaryotic analogue
The complexity in bacterial responses appears to be in the number of proteins induced by oxidative stress. In mammalian cells, the number of proteins induced is small but the regulatory pathways are highly complex.
The inducers of oxidative stress responses in bacteria appear to be either the oxidant itself or interaction of the oxidant with a cell component. Most mammalian cells exist in an environment where the oxygen concentration is constant, thus responses are not directly stimulated by oxidants. Rather, cytokines such as tumor necrosis factor, interleukin-1 or bacterial polysaccharides induce SOD synthesis and multigene responses. Recent work shows that superoxide is a strong tumor promoter that works by activation and induction growth-competence related gene products. Other factors involved in the antioxidant gene expression include an induction of calmodulin kinase by increase in Ca2+ concentrations.
E. coli cells have revealed similarities to the aging process of higher organisms. The similarities include increased oxidation of cellular constituents and its target specificity, the role of antioxidants and oxygen tension in determining life span, and an apparent trade-off between activities related to reproduction and survival.[5]
References
- Semchyshyn, Halyna (2009). "Hydrogen peroxide-induced response in E. coli and S. cerevisiae: different stages of the flow of the genetic information". Open Life Sciences. 4 (2): 142–153. doi:10.2478/s11535-009-0005-5.
- Farr, SB; Kogoma, T (1991). "Oxidative stress responses in Escherichia coli and Salmonella typhimurium". Microbiol Rev. 55 (4): 561–85. doi:10.1128/mr.55.4.561-585.1991. PMC 372838. PMID 1779927.
- Nunoshiba, T; Hidalgo, E; Li, Z; Demple, B (1993). "Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response". J Bacteriol. 175 (22): 7492–4. doi:10.1128/jb.175.22.7492-7494.1993. PMC 206898. PMID 8226698.
- Cabiscol, E; Tamarit, J; Ros, J (2000). "Oxidative stress in bacteria and protein damage by reactive oxygen species". Int Microbiol. 3 (1): 3–8. PMID 10963327.
- Thomas Nystrom, STATIONARY-PHASE PHYSIOLOGY, Annu. Rev. Microbiol. 2004. 58:161–81.