Morpholino
A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule (colloquially, an oligo) used in molecular biology to modify gene expression. Its molecular structure contains DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups. Morpholinos block access of other molecules to small (~25 base) specific sequences of the base-pairing surfaces of ribonucleic acid (RNA). Morpholinos are used as research tools for reverse genetics by knocking down gene function.
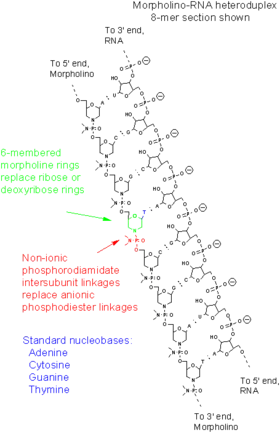
This article discusses only the Morpholino antisense oligomers, which are nucleic acid analogs. The word "Morpholino" can occur in other chemical names, referring to chemicals containing a six-membered morpholine ring. To help avoid confusion with other morpholine-containing molecules, when describing oligos "Morpholino" is often capitalized as a trade name, but this usage is not consistent across scientific literature. Morpholino oligos are sometimes referred to as PMO (for phosphorodiamidate morpholino oligomer), especially in medical literature. Vivo-Morpholinos and PPMO are modified forms of Morpholinos with chemical groups covalently attached to facilitate entry into cells.
Gene knockdown is achieved by reducing the expression of a particular gene in a cell. In the case of protein-coding genes, this usually leads to a reduction in the quantity of the corresponding protein in the cell. Knocking down gene expression is a method for learning about the function of a particular protein; in a similar manner, causing a specific exon to be spliced out of the RNA transcript encoding a protein can help to determine the function of the protein moiety encoded by that exon or can sometimes knock down the protein activity altogether. These molecules have been applied to studies in several model organisms, including mice, zebrafish, frogs and sea urchins.[1] Morpholinos can also modify the splicing of pre-mRNA[2] or inhibit the maturation and activity of miRNA.[3] Techniques for targeting Morpholinos to RNAs and delivering Morpholinos into cells have recently been reviewed in a journal article[4] and in book form.[5]
Morpholinos are in development as pharmaceutical therapeutics targeted against pathogenic organisms such as bacteria[6] or viruses[7] and genetic diseases.[8] A Morpholino-based drug eteplirsen from Sarepta Therapeutics received accelerated approval from the US Food and Drug Administration in September 2016 for the treatment of some mutations causing Duchenne muscular dystrophy,[9] although the approval process was mired in controversy. Other Morpholino-based drugs golodirsen, viltolarsen, and casimersen (also for Duchenne muscular dystrophy) were approved by the FDA in 2019–2021.[10][11][12]
History
Morpholino oligos were conceived by Summerton (Gene Tools) at AntiVirals Inc. (now Sarepta Therapeutics) and originally developed in collaboration with Weller.[13]
Structure
Morpholinos are synthetic molecules that are the product of a redesign of natural nucleic acid structure.[14] Usually 25 bases in length, they bind to complementary sequences of RNA or single-stranded DNA by standard nucleic acid base-pairing. In terms of structure, the difference between Morpholinos and DNA is that, while Morpholinos have standard nucleic acid bases, those bases are bound to methylenemorpholine rings linked through phosphorodiamidate groups instead of phosphates.[14] The figure compares the structures of the two strands depicted there, one of RNA and the other of a Morpholino. Replacement of anionic phosphates with the uncharged phosphorodiamidate groups eliminates ionization in the usual physiological pH range, so Morpholinos in organisms or cells are uncharged molecules. The entire backbone of a Morpholino is made from these modified subunits.
Function
Morpholinos do not trigger the degradation of their target RNA molecules, unlike many antisense structural types (e.g., phosphorothioates, siRNA). Instead, Morpholinos act by "steric blocking", binding to a target sequence within an RNA, inhibiting molecules that might otherwise interact with the RNA.[15] Morpholino oligos are often used to investigate the role of a specific mRNA transcript in an embryo. Developmental biologists inject Morpholino oligos into eggs or embryos of zebrafish,[16] African clawed frog (Xenopus),[17] sea urchin[18] and killifish (F. heteroclitus) producing morphant embryos, or electroporate Morpholinos into chick[19] embryos at later development stages. With appropriate cytosolic delivery systems, Morpholinos are effective in cell culture.[20][21] Vivo-Morpholinos, in which the oligo is covalently linked to a delivery dendrimer, enter cells when administered systemically in adult animals or in tissue cultures.[22]
Normal gene expression in eukaryotes
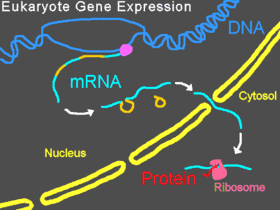
In eukaryotic organisms, pre-mRNA is transcribed in the nucleus, introns are spliced out, then the mature mRNA is exported from the nucleus to the cytoplasm. The small subunit of the ribosome usually starts by binding at the 5' end of the mRNA and is joined there by various other eukaryotic initiation factors, forming the initiation complex. The initiation complex scans along the mRNA strand until it reaches a start codon, and then the large subunit of the ribosome attaches to the small subunit and translation of a protein begins. This entire process is referred to as gene expression; it is the process by which the information in a gene, encoded as a sequence of bases in DNA, is converted into the structure of a protein. A Morpholino can modify splicing, block translation, or block other functional sites on RNA depending on the Morpholino's base sequence.
Blocking translation
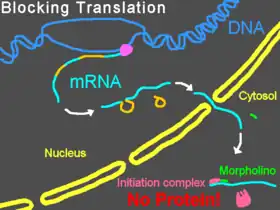
Bound to the 5'-untranslated region of messenger RNA (mRNA), Morpholinos can interfere with progression of the ribosomal initiation complex from the 5' cap to the start codon. This prevents translation of the coding region of the targeted transcript (called "knocking down" gene expression). This is useful experimentally when an investigator wishes to know the function of a particular protein; Morpholinos provide a convenient means of knocking down expression of the protein and learning how that knockdown changes the cells or organism. Some Morpholinos knock down expression so effectively that, after degradation of preexisting proteins, the targeted proteins become undetectable by Western blot.
In 2016 a synthetic peptide-conjugated PMO (PPMO) was found to inhibit the expression of New Delhi Metallo-beta-lactamase, an enzyme that many drug-resistant bacteria use to destroy carbapenems.[23][24]
Modifying pre-mRNA splicing
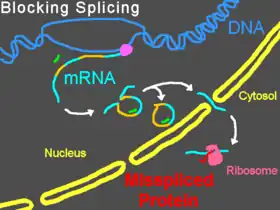
Morpholinos can interfere with pre-mRNA processing steps either by preventing splice-directing small nuclear ribonucleoproteins (snRNP) complexes from binding to their targets at the borders of introns on a strand of pre-mRNA, or by blocking the nucleophilic adenine base and preventing it from forming the splice lariat structure, or by interfering with the binding of splice regulatory proteins such as splice silencers[25] and splice enhancers.[26] Preventing the binding of snRNP U1 (at the donor site) or U2/U5 (at the polypyrimidine moiety and acceptor site) can cause modified splicing, commonly excluding exons from the mature mRNA. Targeting some splice targets results in intron inclusions, while activation of cryptic splice sites can lead to partial inclusions or exclusions.[27] Targets of U11/U12 snRNPs can also be blocked.[28] Splice modification can be conveniently assayed by reverse-transcriptase polymerase chain reaction (RT-PCR) and is seen as a band shift after gel electrophoresis of RT-PCR products.[2]
Other applications: blocking other mRNA sites and use as probes
Morpholinos have been used to block miRNA activity[29][30] and maturation.[3] Fluorescein-tagged Morpholinos combined with fluorescein-specific antibodies can be used as probes for in-situ hybridization to miRNAs.[31] Morpholinos can block ribozyme activity.[32] U2 and U12 snRNP functions have been inhibited by Morpholinos.[33] Morpholinos targeted to "slippery" mRNA sequences within protein coding regions can induce translational frameshifts.[34] Morpholinos can block RNA editing,[35] poly-A tailing[36] and translocation sequences.[37] Morpholino activities against this variety of targets suggest that Morpholinos can be used as a general-purpose tool for blocking interactions of proteins or nucleic acids with mRNA.
Specificity, stability and non-antisense effects
Morpholinos have become a standard knockdown tool in animal embryonic systems, which have a broader range of gene expression than adult cells and can be strongly affected by an off-target interaction. Following initial injections into frog or fish embryos at the single-cell or few-cell stages, Morpholino effects can be measured up to five days later,[38] after most of the processes of organogenesis and differentiation are past, with observed phenotypes consistent with target-gene knockdown. Control oligos with irrelevant sequences usually produce no change in embryonic phenotype, evidence of the Morpholino oligo's sequence-specificity and lack of non-antisense effects. The dose required for a knockdown can be reduced by coinjection of several Morpholino oligos targeting the same mRNA, which is an effective strategy for reducing or eliminating dose-dependent off-target RNA interactions.[39]
mRNA rescue experiments can sometimes restore the wild-type phenotype to the embryos and provide evidence for the specificity of a Morpholino. In an mRNA rescue, a Morpholino is co-injected with an mRNA that codes for the morphlino's protein. However, the rescue mRNA has a modified 5'-UTR (untranslated region) so that the rescue mRNA contains no target for the Morpholino. The rescue mRNA's coding region encodes the protein of interest. Translation of the rescue mRNA replaces production of the protein that was knocked down by the Morpholino. Since the rescue mRNA would not affect phenotypic changes due to the Morpholino's off-target gene expression modulation, this return to wild-type phenotype is further evidence of Morpholino specificity.[38] In some cases, ectopic expression of the rescue RNA makes recovery of the wild-type phenotype impossible.
In embryos, Morpholinos can be tested in null mutants to check for unexpected RNA interactions, then used in a wild-type embryo to reveal the acute knockdown phenotype. The knockdown phenotype is often more extreme than the mutant phenotype; in the mutant, effects of losing the null gene can be concealed by genetic compensation.[40]
Because of their completely unnatural backbones, Morpholinos are not recognized by cellular proteins. Nucleases do not degrade Morpholinos,[41] nor are they degraded in serum or in cells.[42]
Up to 18% of Morpholinos appear to induce nontarget-related phenotypes including cell death in the central nervous system and somite tissues of zebrafish embryos.[43] Most of these effects are due to activation of p53-mediated apoptosis and can be suppressed by co-injection of an anti-p53 Morpholino along with the experimental Morpholino. Moreover, the p53-mediated apoptotic effect of a Morpholino knockdown has been phenocopied using another antisense structural type, showing the p53-mediated apoptosis to be a consequence of the loss of the targeted protein and not a consequence of the knockdown oligo type.[44] It appears that these effects are sequence-specific; as in most cases, if a Morpholino is associated with non-target effects, the 4-base mismatch Morpholino will not trigger these effects.
A cause for concern in the use of Morpholinos is the potential for "off-target" effects. Whether an observed morphant phenotype is due to the intended knockdown or an interaction with an off-target RNA can often be addressed in embryos by running another experiment to confirm that the observed morphant phenotype results from the knockdown of the expected target. This can be done by recapitulating the morphant phenotype with a second, non-overlapping Morpholino targeting the same mRNA,[38] by confirmation of the observed phenotypes by comparing with a mutant strain (though compensation will obscure a phenotype in some mutants), by testing the Morpholino in a null mutant background to detect additional phenotypic changes or by dominant-negative methods. As mentioned above, rescue of observed phenotypes by coinjecting a rescue mRNA is, when feasible, a reliable test of specificity of a Morpholino.[38][40]
Delivery
For a Morpholino to be effective, it must be delivered past the cell membrane into the cytosol of a cell. Once in the cytosol, Morpholinos freely diffuse between the cytosol and nucleus, as demonstrated by the nuclear splice-modifying activity of Morpholinos observed after microinjection into the cytosol of cells. Different methods are used for delivery into embryos, into cultured cells or into adult animals. A microinjection apparatus is usually used for delivery into an embryo, with injections most commonly performed at the single-cell or few-cell stage;[45] an alternative method for embryonic delivery is electroporation, which can deliver oligos into tissues of later embryonic stages.[46] Common techniques for delivery into cultured cells include the Endo-Porter peptide (which causes the Morpholino to be released from endosomes),[21] the Special Delivery system (no longer commercially available, used a Morpholino-DNA heteroduplex and an ethoxylated polyethylenimine delivery reagent),[20] electroporation,[47] or scrape loading.[48]
Delivery into adult tissues is usually difficult, though there are a few systems allowing useful uptake of unmodified Morpholino oligos (including uptake into muscle cells with Duchenne muscular dystrophy[49] or the vascular endothelial cells stressed during balloon angioplasty[50]). Though they permeate through intercellular spaces in tissues effectively, unconjugated PMOs have limited distribution into the cytosol and nuclear spaces within healthy tissues following IV administration. Systemic delivery into many cells in adult organisms can be accomplished by using covalent conjugates of Morpholino oligos with cell-penetrating peptides, and, while toxicity has been associated with moderate doses of the peptide conjugates,[51][52] they have been used in vivo for effective oligo delivery at doses below those causing observed toxicity.[7][53] An octa-guanidinium dendrimer attached to the end of a Morpholino can deliver the modified oligo (called a Vivo-Morpholino) from the blood to the cytosol.[22][54] Delivery-enabled Morpholinos, such as peptide conjugates and Vivo-Morpholinos, show promise as therapeutics for viral and genetic diseases.[55]
References
- Heasman J (March 2002). "Morpholino oligos: making sense of antisense?". Developmental Biology. 243 (2): 209–14. doi:10.1006/dbio.2001.0565. PMID 11884031.
- Draper BW, Morcos PA, Kimmel CB (July 2001). "Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown". Genesis. 30 (3): 154–6. doi:10.1002/gene.1053. PMID 11477696.
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH (August 2007). "Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development". PLOS Biology. 5 (8): e203. doi:10.1371/journal.pbio.0050203. PMC 1925136. PMID 17676975.
- Moulton JD (2016). "Guide for Morpholino Users: Toward Therapeutics" (PDF). J Drug Discov Develop and Deliv. 3 (2): 1023.
- Moulton HM, Moulton JD, eds. (2017). Morpholino Oligomers. Methods in Molecular Biology. Vol. 1565. Humana Press (Springer). p. 284. doi:10.1007/978-1-4939-6817-6. ISBN 978-1-4939-6815-2. S2CID 28338321.
- Geller BL (April 2005). "Antibacterial antisense". Current Opinion in Molecular Therapeutics. 7 (2): 109–13. PMID 15844617.
- Deas TS, Bennett CJ, Jones SA, Tilgner M, Ren P, Behr MJ, Stein DA, Iversen PL, Kramer LD, Bernard KA, Shi PY (July 2007). "In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus". Antimicrobial Agents and Chemotherapy. 51 (7): 2470–82. doi:10.1128/AAC.00069-07. PMC 1913242. PMID 17485503.
- McClorey G, Fall AM, Moulton HM, Iversen PL, Rasko JE, Ryan M, Fletcher S, Wilton SD (October 2006). "Induced dystrophin exon skipping in human muscle explants". Neuromuscular Disorders. 16 (9–10): 583–90. doi:10.1016/j.nmd.2006.05.017. PMID 16919955. S2CID 21338534.
- "Press Announcements - FDA grants accelerated approval to first drug for Duchenne muscular dystrophy". Food and Drug Administration. 2019-09-10.
- Commissioner, Office of the (2019-12-12). "FDA grants accelerated approval to first targeted treatment for rare Duchenne muscular dystrophy mutation". FDA. Retrieved 2019-12-14.
- Roshmi, R. R.; Yokota, T. (October 2019). "Viltolarsen for the treatment of Duchenne muscular dystrophy". Drugs of Today. 55 (10): 627–639. doi:10.1358/dot.2019.55.10.3045038. ISSN 1699-3993. PMID 31720560. S2CID 207935659.
- Shirley, Matt (May 2021). "Casimersen: First Approval". Drugs. 81 (7): 875–879. doi:10.1007/s40265-021-01512-2. ISSN 1179-1950. PMID 33861387. S2CID 233248050.
- Summerton JE (2017). "Invention and Early History of Morpholinos: From Pipe Dream to Practical Products". Morpholino Oligomers. Methods in Molecular Biology. Vol. 1565. Humana Press (Springer). pp. 1–15. doi:10.1007/978-1-4939-6817-6_1. ISBN 978-1-4939-6817-6. PMID 28364229.
- Summerton J, Weller D (June 1997). "Morpholino antisense oligomers: design, preparation, and properties". Antisense & Nucleic Acid Drug Development. 7 (3): 187–95. doi:10.1089/oli.1.1997.7.187. PMID 9212909. S2CID 19372403.
- Summerton J (December 1999). "Morpholino antisense oligomers: the case for an RNase H-independent structural type". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1489 (1): 141–58. doi:10.1016/S0167-4781(99)00150-5. PMID 10807004.
- Nasevicius A, Ekker SC (October 2000). "Effective targeted gene 'knockdown' in zebrafish". Nature Genetics. 26 (2): 216–20. doi:10.1038/79951. PMID 11017081. S2CID 21451111.
- Heasman J, Kofron M, Wylie C (June 2000). "Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach". Developmental Biology. 222 (1): 124–34. doi:10.1006/dbio.2000.9720. PMID 10885751.
- Howard EW, Newman LA, Oleksyn DW, Angerer RC, Angerer LM (February 2001). "SpKrl: a direct target of beta-catenin regulation required for endoderm differentiation in sea urchin embryos". Development. 128 (3): 365–75. doi:10.1242/dev.128.3.365. PMID 11152635.
- Kos R, Reedy MV, Johnson RL, Erickson CA (April 2001). "The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos". Development. 128 (8): 1467–79. doi:10.1242/dev.128.8.1467. PMID 11262245.
- Morcos PA (July 2001). "Achieving efficient delivery of morpholino oligos in cultured cells". Genesis. 30 (3): 94–102. doi:10.1002/gene.1039. PMID 11477682.
- Summerton JE (November 2005). "Endo-Porter: a novel reagent for safe, effective delivery of substances into cells". Annals of the New York Academy of Sciences. 1058 (1): 62–75. Bibcode:2005NYASA1058...62S. doi:10.1196/annals.1359.012. PMID 16394126. S2CID 28590429.
- Morcos PA, Li Y, Jiang S (December 2008). "Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues". BioTechniques. 45 (6): 613–4, 616, 618 passim. doi:10.2144/000113005. PMID 19238792.
- "New molecule knocks out superbugs' immunity to antibiotics". newatlas.com. 2017-01-20. Retrieved 2017-01-25.
- Sully EK, Geller BL, Li L, Moody CM, Bailey SM, Moore AL, Wong M, Nordmann P, Daly SM, Sturge CR, Greenberg DE (March 2017). "Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo". The Journal of Antimicrobial Chemotherapy. 72 (3): 782–790. doi:10.1093/jac/dkw476. PMC 5890718. PMID 27999041.
- Bruno IG, Jin W, Cote GJ (October 2004). "Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements". Human Molecular Genetics. 13 (20): 2409–20. doi:10.1093/hmg/ddh272. PMID 15333583.
- Vetrini F, Tammaro R, Bondanza S, Surace EM, Auricchio A, De Luca M, Ballabio A, Marigo V (May 2006). "Aberrant splicing in the ocular albinism type 1 gene (OA1/GPR143) is corrected in vitro by morpholino antisense oligonucleotides". Human Mutation. 27 (5): 420–6. doi:10.1002/humu.20303. PMID 16550551. S2CID 21092974.
- Morcos PA (June 2007). "Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos". Biochemical and Biophysical Research Communications. 358 (2): 521–7. doi:10.1016/j.bbrc.2007.04.172. PMID 17493584.
- König H, Matter N, Bader R, Thiele W, Müller F (November 2007). "Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation". Cell. 131 (4): 718–29. doi:10.1016/j.cell.2007.09.043. PMID 18022366.
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH (2004). "Substrate requirements for let-7 function in the developing zebrafish embryo". Nucleic Acids Research. 32 (21): 6284–91. doi:10.1093/nar/gkh968. PMC 535676. PMID 15585662.
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (February 2007). "Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate". Nature Genetics. 39 (2): 259–63. doi:10.1038/ng1953. PMC 3982799. PMID 17220889.
- Lagendijk AK, Moulton JD, Bakkers J (June 2012). "Revealing details: whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues". Biology Open. 1 (6): 566–9. doi:10.1242/bio.2012810. PMC 3509442. PMID 23213449.
- Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC (September 2004). "Exogenous control of mammalian gene expression through modulation of RNA self-cleavage". Nature. 431 (7007): 471–6. Bibcode:2004Natur.431..471Y. doi:10.1038/nature02844. PMID 15386015. S2CID 4425795.
- Matter N, König H (February 2005). "Targeted 'knockdown' of spliceosome function in mammalian cells". Nucleic Acids Research. 33 (4): e41. doi:10.1093/nar/gni041. PMC 549580. PMID 15731334.
- Howard MT, Gesteland RF, Atkins JF (October 2004). "Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides". RNA. 10 (10): 1653–61. doi:10.1261/rna.7810204. PMC 1370650. PMID 15383681.
- Penn AC, Balik A, Greger IH (January 2013). "Steric antisense inhibition of AMPA receptor Q/R editing reveals tight coupling to intronic editing sites and splicing". Nucleic Acids Research. 41 (2): 1113–23. doi:10.1093/nar/gks1044. PMC 3553965. PMID 23172291.
- Vorlová S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L (September 2011). "Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation". Molecular Cell. 43 (6): 927–39. doi:10.1016/j.molcel.2011.08.009. PMC 3781938. PMID 21925381.
- Arthur PK, Claussen M, Koch S, Tarbashevich K, Jahn O, Pieler T (July 2009). "Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis". The Journal of Biological Chemistry. 284 (30): 19982–92. doi:10.1074/jbc.M109.009928. PMC 2740424. PMID 19458392.
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC (March 2009). "A primer for morpholino use in zebrafish". Zebrafish. 6 (1): 69–77. doi:10.1089/zeb.2008.0555. PMC 2776066. PMID 19374550.
- Kamachi Y, Okuda Y, Kondoh H (January 2008). "Quantitative assessment of the knockdown efficiency of morpholino antisense oligonucleotides in zebrafish embryos using a luciferase assay". Genesis. 46 (1): 1–7. doi:10.1002/dvg.20361. PMID 18196596.
- Stainier DY, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, Ingham PW, Schulte-Merker S, Yelon D, Weinstein BM, Mullins MC, Wilson SW, Ramakrishnan L, Amacher SL, Neuhauss SC, Meng A, Mochizuki N, Panula P, Moens CB (October 2017). "Guidelines for morpholino use in zebrafish". PLOS Genetics. 13 (10): e1007000. doi:10.1371/journal.pgen.1007000. PMC 5648102. PMID 29049395.
- Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD (1996). "Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation". Antisense & Nucleic Acid Drug Development. 6 (4): 267–72. doi:10.1089/oli.1.1996.6.267. PMID 9012862.
- Youngblood DS, Hatlevig SA, Hassinger JN, Iversen PL, Moulton HM (2007). "Stability of cell-penetrating peptide-morpholino oligomer conjugates in human serum and in cells". Bioconjugate Chemistry. 18 (1): 50–60. doi:10.1021/bc060138s. PMID 17226957.
- Ekker SC, Larson JD (July 2001). "Morphant technology in model developmental systems". Genesis. 30 (3): 89–93. doi:10.1002/gene.1038. PMID 11477681.
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC (May 2007). "p53 activation by knockdown technologies". PLOS Genetics. 3 (5): e78. doi:10.1371/journal.pgen.0030078. PMC 1877875. PMID 17530925.
- Rosen JN, Sweeney MF, Mably JD (March 2009). "Microinjection of zebrafish embryos to analyze gene function". Journal of Visualized Experiments. 9 (25). doi:10.3791/1115. PMC 2762901. PMID 19274045.
- Cerda GA, Thomas JE, Allende ML, Karlstrom RO, Palma V (July 2006). "Electroporation of DNA, RNA, and morpholinos into zebrafish embryos". Methods. 39 (3): 207–11. doi:10.1016/j.ymeth.2005.12.009. hdl:10533/178089. PMID 16837210.
- Jubin R (2004). "Optimizing electroporation conditions for intracellular delivery of Morpholino antisense oligonucleotides directed against the hepatitis C virus internal ribosome entry site". Antisense Therapeutics. pp. 309–22. doi:10.1385/1-59259-854-4:309. ISBN 978-1-59259-854-0. PMID 15375324.
{{cite book}}:|journal=ignored (help) - Partridge M, Vincent A, Matthews P, Puma J, Stein D, Summerton J (1996). "A simple method for delivering morpholino antisense oligos into the cytoplasm of cells". Antisense & Nucleic Acid Drug Development. 6 (3): 169–75. doi:10.1089/oli.1.1996.6.169. PMID 8915501. S2CID 8161222.
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD, Wilton SD (February 2006). "Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide". The Journal of Gene Medicine. 8 (2): 207–16. doi:10.1002/jgm.838. PMID 16285002. S2CID 42463260.
- Kipshidze NN, Kim HS, Iversen P, Yazdi HA, Bhargava B, New G, Mehran R, Tio F, Haudenschild C, Dangas G, Stone GW, Iyer S, Roubin GS, Leon MB, Moses JW (May 2002). "Intramural coronary delivery of advanced antisense oligonucleotides reduces neointimal formation in the porcine stent restenosis model". Journal of the American College of Cardiology. 39 (10): 1686–91. doi:10.1016/S0735-1097(02)01830-2. PMID 12020498.
- Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL, Lebleu B (December 2006). "Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents". Journal of Controlled Release. 116 (3): 304–13. doi:10.1016/j.jconrel.2006.09.011. PMID 17097177.
- Burrer R, Neuman BW, Ting JP, Stein DA, Moulton HM, Iversen PL, Kuhn P, Buchmeier MJ (June 2007). "Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models". Journal of Virology. 81 (11): 5637–48. doi:10.1128/JVI.02360-06. PMC 1900280. PMID 17344287.
- Amantana A, Moulton HM, Cate ML, Reddy MT, Whitehead T, Hassinger JN, Youngblood DS, Iversen PL (2007). "Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate". Bioconjugate Chemistry. 18 (4): 1325–31. doi:10.1021/bc070060v. PMID 17583927.
- Li YF, Morcos PA (July 2008). "Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo". Bioconjugate Chemistry. 19 (7): 1464–70. doi:10.1021/bc8001437. PMID 18564870.
- Moulton JD, Jiang S (March 2009). "Gene knockdowns in adult animals: PPMOs and vivo-morpholinos". Molecules. 14 (3): 1304–23. doi:10.3390/molecules14031304. PMC 6253989. PMID 19325525.
Further reading
- Wiley-Liss, Inc. Special Issue: Morpholino Gene Knockdowns of genesis Volume 30, Issue 3 Pages 89-200 (July 2001). This is a special issue of Genesis that consists of a series of peer-reviewed short papers using Morpholino knock downs of gene function in various animal and tissue culture systems.
- "Peptide Nucleic Acids, Morpholinos and Related Antisense Biomolecules." eds. Janson & During (Springer, 2007)
- Moulton J (2007). "Using Morpholinos to Control Gene Expression (Unit 4.30)". In Beaucage S (ed.). Current Protocols in Nucleic Acid Chemistry. New Jersey: John Wiley & Sons, Inc. ISBN 978-0-471-24662-6.