Pii nitrogen regulatory proteins
The PII family comprises a group of widely distributed signal transduction proteins found in nearly all Bacteria and also present in Archaea and in the chloroplasts of Algae and plants.[2] PII form barrel-like homotrimers with a flexible loop, namely T-loop, emerging from each subunit.[2] PII proteins have extraordinary sensory properties; they can exist in a vast range of structural status accordingly to the levels of ATP, ADP and 2-oxogluratate. These metabolites interact allosterically with PII in three conserved binding sites located in the lateral cavity between each PII subunit. ATP and ADP bind competitively to the nucleotide binding whereas the 2-oxoglutarate only interacts with PII in the presence of MgATP.[3]
| Nitrogen regulatory protein P-II | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | P-II | ||||||||
| Pfam | PF00543 | ||||||||
| Pfam clan | CL0089 | ||||||||
| InterPro | IPR002187 | ||||||||
| SMART | SM00938 | ||||||||
| PROSITE | PDOC00439 | ||||||||
| SCOP2 | 1pil / SCOPe / SUPFAM | ||||||||
| |||||||||
In Proteobacteria, PII proteins are also subject to a cycle of reversible posttranslational modification (Huergo et al., 2013). Under a low nitrogen regime, the low intracellular glutamine level triggers the uridylyl-transferase activity of the bi-functional GlnD enzyme promoting the uridylylation of a conserved Tyr-51 located at the top the PII T-loop. Conversely, under a high nitrogen regime, accumulation of intracellular glutamine triggers the uridylyl-removing activity of GlnD and PII accumulates in its non-modified form (Huergo et al., 2013).
The ability of PII to sense important metabolites and integrate signals deriving from energy status (ATP and ADP ratio), carbon (2-oxogluratate) and nitrogen (glutamine and 2-oxoglutarate) levels were capitalized during evolution such that PII can act as a dissociable regulatory subunit of a range of transporters, transcriptional regulators and enzymes.[4]
Structure
PII proteins exist in trimers in vivo and bind ATP in a cleft between the subunits. There are two flexible loops call the B-loop and T-loop which are involved in regulation of the protein. The T-loop contains a conserved tyrosine which is the site of uridyl attachment.
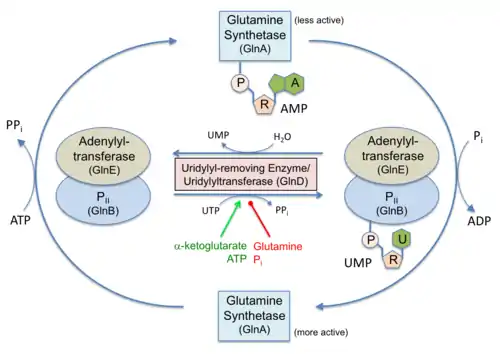
Role in nitrogen metabolism
Following nitrogen starvation, increased intra-cellular concentrations of ammonia cause the de-uridylylation of GlnK. This then binds directly to the ammonia channel AmtB to block ammonia conduction.[5][6] PII proteins such as SbtB are also implicated in carbon metabolism regulation, these proteins are able to control the activity of Acetyl-CoA carboxylase in plants, algae and Bacteria [7]
References
- Carr PD, Cheah E, Suffolk PM, Vasudevan SG, Dixon NE, Ollis DL (January 1996). "X-ray structure of the signal transduction protein from Escherichia coli at 1.9 A". Acta Crystallographica Section D. 52 (Pt 1): 93–104. doi:10.1107/S0907444995007293. PMID 15299730.
- Huergo LF, Chandra G, Merrick M (March 2013). "P(II) signal transduction proteins: nitrogen regulation and beyond". FEMS Microbiology Reviews. 37 (2): 251–83. doi:10.1111/j.1574-6976.2012.00351.x. PMID 22861350.
- Huergo LF, Pedrosa FO, Muller-Santos M, Chubatsu LS, Monteiro RA, Merrick M, Souza EM (January 2012). "PII signal transduction proteins: pivotal players in post-translational control of nitrogenase activity". Microbiology. 158 (Pt 1): 176–90. doi:10.1099/mic.0.049783-0. PMID 22210804.
- Huergo LF, Dixon R (December 2015). "The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite". Microbiology and Molecular Biology Reviews. 79 (4): 419–35. doi:10.1128/MMBR.00038-15. PMC 4651028. PMID 26424716.
- Durand A, Merrick M (October 2006). "In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate". The Journal of Biological Chemistry. 281 (40): 29558–67. doi:10.1074/jbc.M602477200. PMID 16864585.
- Conroy MJ, Durand A, Lupo D, Li XD, Bullough PA, Winkler FK, Merrick M (January 2007). "The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel". Proceedings of the National Academy of Sciences of the United States of America. 104 (4): 1213–8. Bibcode:2007PNAS..104.1213C. doi:10.1073/pnas.0610348104. PMC 1783118. PMID 17220269.
- Gerhardt EC, Rodrigues TE, Müller-Santos M, Pedrosa FO, Souza EM, Forchhammer K, Huergo LF (March 2015). "The bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase". Molecular Microbiology. 95 (6): 1025–35. doi:10.1111/mmi.12912. PMID 25557370. S2CID 20384007.