Polaribacter
Polaribacter is a genus in the family Flavobacteriaceae. They are gram-negative, aerobic bacteria that can be heterotrophic, psychrophilic or mesophilic.[2] Most species are non-motile and species range from ovoid to rod-shaped.[2] Polaribacter forms yellow- to orange-pigmented colonies.[2] They have been mostly adapted to cool marine ecosystems, and their optimal growth range is at a temperature between 10 and 32 °C and at a pH of 7.0 to 8.0.[2][3] They are oxidase and catalase-positive and are able to grow using carbohydrates, amino acids, and organic acids.[2]
| Polaribacter | |
|---|---|
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Polaribacter Gosink et al. 1998[1] |
| Type species | |
| Polaribacter filamentus[1] | |
| Species | |
|
P. aestuariivivens[1] | |
There is evidence of two life strategies for members of the genus, Polaribacter. Some Polaribacter species are free-living and consume amino acids and carbohydrates, as well as have proteorhodopsin that enhances living in oligotrophic seawaters.[4] Other species of Polaribacter attach to substrates in search of protein polymers.[4]
In the context of climate change, algal blooms are becoming increasingly prevalent.[5] Members of the genus Polaribacter decompose algal cells and thus may be important in biogeochemical cycling, as well as influence seawater chemistry and the composition of microbial communities as temperatures continue to rise. This may impact the efficiency of the biological pump in sequestering atmospheric carbon.[6]
Polaribacter is a genus that is being continuously researched and to date there are 25 species that have been validly published under the International Code of Nomenclature of Prokaryotes (ICNP): P. aquimarinus, P. atrinae, P. butkevichii, P. dokdonensis, P. filamentus, P. franzmannii, P. gangjinensis, P. glomeratus, P. haliotis, P. huanghezhanensis, P. insulae, P. irgensii, P. lacunae, P. litorisediminis, P. marinaquae, P. marinivivus, P. pacificus, P. porphyrae, P. reichenbachii, P. sejongensis, P. septentrionalilitoris, P. staleyi, P. tangerinus, P. undariae, P. vadi.
The genus is sometimes incorrectly referred to as Polaribacer; Polarobacter or Polaribacteria.[7]
Phylogeny
This phylogeny is based on rRNA gene sequencing.[8]
| Polaribacter |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| outgroup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Distribution and Abundance
Members in the genus Polaribacter are abundant in polar oceans and are important in the export of dissolved organic matter (DOM).[9][10] A small percentage of the bacterial community is responsible for the DOM uptake rate.[11]
In northern latitude waters, the fraction of cells using glucose (fraction of active cells) is higher in summer than winter,[11] and high abundances may occur after phytoplankton blooms,[12][13] although a study in southern high-latitude waters found lower abundances of Polaribacter after an in situ diatom bloom.[14]
Within the Arctic Ocean, there is no obvious pattern in the relative abundance between summer and winter.[11] In the Chukchi Sea, the fraction of cells using leucine is higher in the winter than in summer.[11] In the Beaufort Sea, the fraction of cells using leucine does not differ between seasons.[11] In the coastal waters of Fildes Peninsula, Polaribacter dominated cells in the phylum Bacteriodetes.[15]
Habitat

Microorganisms in the genus Polaribacter are widely distributed and various species are capable of living in a plethora of different environments. Some Polaribacter species have been isolated from brine pools in the Arctic Ocean.[16] in addition to hypersaline environments, numerous Polaribacter species inhabit extreme environments ranging from -20 °C to 22 °C.[17] In the past, it was thought that Polaribacter only flourished in cold waters as the members of the species that were first discovered (P. irgensii, P. filamentus, and P. franzmannii) in the Arctic and Southern Oceans could only survive in water with temperatures ranging from -20 °C to 10 °C.[17][18] Subsequently, members of the genus Polaribacter have been shown to be very versatile microorganisms and can survive in oligotrophic and in copiotrophic environments.[17] Polaribacter have also been found in sediments.[19] For example, SM1202T, a phylogenetically close strain to Polaribacter was isolated from marine sediment in Kongsfjorden, Svalbard.[19] Polaribacter have also been experimentally isolated from red macroalgae (Porphyra yezoensis) and green macroalgae (Ulva fenestrate).[20][21]
Role in ecosystem
Isolates of related Flavobacteria are able to degrade High-Molecular Weight (HMW) DOM.[9] and Polaribacter may be among the first organisms to degrade particulate organic matter and break-down polymers into smaller particles that can be used by free-living bacterial heterotrophs.[22] This suggests that they likely remineralize primary production matter within the food web.[22]
In the Southern Ocean
The Antarctic Peninsula exhibits strong seasonal changes, which influences how bacteria respond to and live in these environmental conditions. The Antarctic spring is especially important as it brings about significant changes, including sea ice melting, thermal stratification due to warming surface waters, and increased dissolved organic matter (DOM) production. All these physical changes also result in phytoplankton blooms which are important in supporting higher trophic levels.[23]
In the Southern Ocean, flavobacteria dominate bacterial activity, particularly flavobacteria in the genus Polaribacter. Typically, these bacteria are prevalent in sea ice; however, during seasonal melting in the summer, they dominate coastal waters as sea ice retreats.[22] In the Southern Ocean, when phytoplankton blooms occur, Flavobacteria, and particularly members in the genus Polaribacter, are among the first bacterial taxa to respond to phytoplankton blooms, breaking down organic matter by direct attachment and the use of exoenzymes.[23][24] Both particle-attached and free-living members of the family Rhodobacteraceae were also found in close association with phytoplankton blooms; however, bacteria in this family were found to use lower molecular weight substrates.[23] This suggests that they're secondary in the microbial succession of substrates, using the byproducts of degradation by flavobacteria, which also includes members of the genus Polaribacter.[23] The relative abundance of free-living bacteria belonging to the genus Polaribacter and in the family Rhodobacteraceae peaked at different points during phytoplankton blooms, suggesting a niche specialization contributing to successive degradation of phytoplankton-derived organic matter.[23] Bacteria in the genus Polaribacter and family Rhodobacteraceae were found in clusters, with Polaribacter clusters forming earlier in the bloom, which further suggests a successive ecological interaction between various bacterial taxa.[23]
For both the Arctic Ocean and the North Sea, Polaribacter exhibited similar trends pertaining to phytoplankton blooms in the summertime as well as assuming particular niches for organic matter degradation.[16]
Metabolism
Members of the genus Polaribacter are metabolically flexible depending on their physiology, lifestyle and seasonality of the region they inhabit.[17] Many research studies have found that Polaribacter can alternate between two lifestyles as a mechanism for adaptation in surface waters where nutrient concentrations are low and light exposure is high.[4] Sequenced strains of the genus Polaribacter show a high prevalence of peptidase and glycoside hydrolase genes in comparison to other bacteria in the Flavobacteriaceae, indicating they contribute to degradation and uptake of external proteins and oligopeptides.[17]
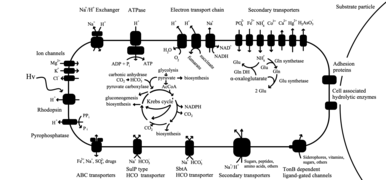
In the pelagic water column, some species are well equipped to attach to particles and substrates to search for and degrade polymers.[25] They are amongst the first organisms to degrade particulate organic matter and break-down polymers into smaller particles.[17] Studies have shown that they will colonize and attach to particles, glide to search for substrates, and degrade them for carbon and nutrients.[17] Once they've degraded these molecules, the bacterium may then search for new particles to colonize, forcing them to freely-swim in environments where nutrients and organic carbon is not easily available.[17]
CAZymes
Genetic sequencing found that strains contain numerous genes which encode for CAZymes that are involved in polysaccharide degradation.[26] For example, strain DSW-5 (a strain genetically very similar to strain MED-152), contains 85 genes encoding to CAZymes and 203 peptidases, which suggests its role as a free-living heterotrophs.[26] However, the ratio of peptidases to glycoside hydrolase genes varies depending on the environmental conditions the strain is subjected to.[17] For example, Polaribacter sp. MED134 lives in environmental conditions with extended starvation conditions and expresses twice as many peptidases as CAZymes.[17] On the other hand, macroalgae-colonizing species that live in stable, eutrophic environments may express greater proportions of CAZymes than peptidases.[17]
Proteorhodopsin
"Free-living" species have the proteorhodopsin gene, which allows them to complete inorganic-carbon fixation using light as an energy source.[4] By utilizing their proteorhodopsin to use light energy, Polaribacter can grow in oligotrophic environmental conditions.[4][17]
Genome
General Genome Characteristics
The genome of bacteria in the genus Polaribacter vary in size from 2.76 Mb (P. irgensii) to 4.10 Mb (P. reichenbachii) and the number of genes ranging from 2446 in P. irgensii to 3500 in P. reichenbachii, but have a fairly constant G+C content of approximately 30 mol%.[27][28] Some notable features of the genome include genes for agar, alginate, and carrageenan degrading enzymes in Polaribacter species which colonize the surface of macroalgae.[27] Agar degrading enzymes have also been found in strains of Polaribacter that colonize the gut of the comb pen shell.[27][29] Proteases are also commonly found in the genomes of species that preferentially grow on solid substrates and degrade protein instead of using free amino acids and living a pelagic lifestyle.[4] Some members of the genus encode proteorhodopsin, which has been implicated in supporting their central metabolism through photophosphorylation.[4]
DNA Sequencing of Polaribacter
DNA sequencing has commonly been used to identify new strains of Polaribacter and help place species on a phylogenetic tree. DNA sequencing has also been used to help understand, or predict a species role in an environment due to the presence of certain genes. Members of the family Flavobacteriaceae can be identified through the specific quinone, Menaquinone 6, also known as Vitamin K2; however, differentiating species can be much more difficult.[28] Species such as Polaribacter vadi and Polaribacter atrinae were identified as new species based on their similar but unique genome when compared to other members of the genus Polaribacter.[29][28] New species can be identified through DNA hybridization or through the sequencing and comparison of a common gene such as 16S rRNA.[28] This has allowed scientists to create phylogenetic trees of the genus based on genomic similarity, as seen in the phylogeny section, as well as identify common features in the genome.[28]
Life Strategies of Polaribacter Based on Genome Analysis
Genomic analysis has allowed scientists to examine the relationships between different species of Polaribacter. However, by combining genomic analysis with other analytical techniques such as chemotaxonomic and biochemical, scientists can theorize how a species might fit into an environment or how they believe a species is adapted to survive.[29][4] A genomic analysis of the Polaribacter strain MED152, found a considerable amount of genes that allow for surface or particle attachment, gliding motility and polymer degradation.[4] These genes fit with the current understanding of how marine bacteroidetes survive through attaching to a surface and moving over it to look for nutrients.[4] However the researchers also noticed that the organism had a proteorhodopsin gene as well as other genes which could be used to sense light and found that under light the species increased carbon dioxide fixation.[4] This led the researchers to theorize that Polaribacter strain MED152 has two different life strategies, one where it acts like other marine bacteroidetes, attaching to surfaces and searching for nutrients and, another life strategy where, if the strain was in a well lit, low nutrient area of the ocean, it would use carbon fixation to synthesize intermediates of metabolic pathways.[4]
Another example of this comes from the Polaribacter strains Hel1_33_49 and Hel1_85. The strain Hel1_33_49 has a genome which contains proteorhodopsin, fewer polysaccharide utilization loci and no mannitol dehydrogenase, which the researchers associate with a pelagic lifestyle.[17] Hel1_85 on the other hand, has a genome which contains twice as many polysaccharide utilization loci, a mannitol dehydrogenase and no proteorhodopsin, pointing to a lifestyle with lower oxygen availability such as a biofilm.[17]
Species
| Name | Type strain[30][1] | DNA G+C content (mol%)[30] | Description[30] |
|---|---|---|---|
| P. aestuariivivens | JDTF-33, KCTC 52838, NBRC 112782[31] | 41.7[31] | Ovoid, coccoid or rod-shaped. Form smooth, glistening, circular, and yellowish-white colonies.[31] |
| P. aquimarinus | ZY113, KCTC 62374, MCCC 1H00296[8] | 30.1[8] | Rod-shaped. Form smooth, circular, and orange colonies[8] |
| P. atrinae | WP2, KACC 17473, JCM 19202 | 30.4 | Rod-shaped, aerobic and non-motile. Form circular, convex, yellow-orange colonies.[29] |
| P. butkevichii | KMM 3938, KCTC 12100, CCUG 48005 | 32.4 | Rod-shaped, mesophillic cells. |
| P. dokdonensis | DSW-5, DSM 17204, KCTC 12392 | 30.0 | Straight or curved rod-shaped. Form smooth, convex, orange colonies. |
| P. filamentus | 215, ATCC 700397, CIP 106479 | 32.0 | Filamentous or rod-shaped. Form gas vesicles. Form orange, flat-convex colonies. |
| P. franzmannii | 301, ATCC 700399, CIP 106480. | 32.5 | Filamentous or rod-shaped. Psychrophilic or psychrotolerant. Form gas vesicles. |
| P. gangjinensis | K17-16, JCM 16152, KCTC 22729 | 34.6 | Gliding motility. Mesophillic. Form smooth, convex, and circular colonies. |
| P. glomeratus | ATCC 43844, CIP 103112, LMG 13858 | 33.0 | Curved or coiled. Psychrophilic or psychrotolerant. |
| P. haliotis | RA4-7, KCTC 52418, NBRC 112383 | 29.9 | Ovoid or rod-shaped. Form smooth, glistening, convex, and light yellow colonies. |
| P. huanghezhanensis | SM1202, CCTCC AB 2013148, KCTC 32516 | 36.4 | Rod-shaped. Form glistening, circular, and orange colonies. |
| P. insulae | OITF-22, KCTC 52658, NBRC 112706 | 32.3 | Ovoid or rod-shaped. Form smooth, glistening, circular, and light orange-yellow colonies. |
| P. irgensii | 23-P, ATCC 700398, CIP 106478 | 34.5 | Filamentous or rod-shaped. Form gas vescicles. Psychrophilic or psychrotolerant. Form translucent, circular, and orange colonies. |
| P. lacunae | HMF2268, KCTC 42191, CECT 8862 | 34.3 | Rod-shaped. Form smooth, circular, and yellow colonies. |
| P. litorisediminis | OITF-11, KCTC 52500, NBRC 112457 | 32.2 | Filamentous, ovoid, or rod-shaped. Form smooth, glistening, circular, and light orange-yellow colonies. |
| P. marinaquae | RZW3-2, JCM 30825, KCTC 42664, MCCC1K00696 | 30.5 | Rod-shaped. Form circular, convex, and yellow colonies. |
| P. marinivivus | GYSW-15, CECT 8655, KCTC 42156 | 31.2 | Rod-shaped. Form smooth, glistening, circular, and yellow colonies. |
| P. pacificus | HRA130-1, KCTC 52370, MCCC 1K03199, JCM31460, CGMCC 1.15763 | 35.9 | Rod-shaped. Form circular, nontransparent, and yellow colonies. |
| P. porphyrae | LNM-20, LMG 26671, NBRC 108759 | 28.6 | Rod-shaped with pointy ends. Lack gas vesicles and polar flagella. Form circular, convex, and pale yellow colonies. |
| P. reichenbachii | 6Alg 8, KCTC 23969, LMG 26443 | 29.1–29.5 | Rod-shaped. Form shiny, circular, and yellow colonies. |
| P. sejongensis | KOPRI 21160, KCTC 23670, JCM 18092 | 29.8 | Rod-shaped. Form circular, convex, and light yellow colonies. |
| P. septentrionalilitoris[32] | ANORD1, DSM 110039, NCIMB 15081, MTCC 12685[32] | 30.6[32] | Cocci or rod-shaped. Form translucent, circular, and bright yellow colonies.[32] |
| P. staleyi | 10Alg 139, KCTC 5277, KMM 6729 | 31.8 | Rod-shaped. Form shiny, circular, and yellow colonies. |
| P. tangerinus | S2-14, KCTC 52275, MCCC 1H00163 | 31.2 | Ovoid or rod-shaped. Form smooth, circular, and orange colonies. |
| P. undariae | W-BA7, KCTC 42175, CECT 8670 | 31.9 | Ovoid or rod-shapted. Form smooth, glistening, circular, and pale yellow colonies. |
| P. vadi | LPB0003, KACC 18704, JCM 31217 | 29.6 | Curved and rod-shaped. Form circular, convex, and yellow colonies.[28] |
Viral pathogens
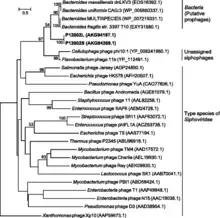
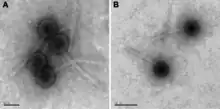
Only two species of lytic phage are known to infect members of this genus, and both have double stranded DNA with virions that include isometric heads and non-contractile tails (class Caudoviricetes, morphotype: siphoviruses).[33] Viral lysis has been implicated as a major driver of changes in genus-level composition of microbial communities.[34]
Applications/uses
Cold water enzymes contained in psychrophilic bacteria like Polaribacter are valuable for biotechnology applications since they do not require high temperatures that may other enzyme systems do.[35]
Psychrophilic enzymes
Polaribacter is a psychrophilic bacterium that lends itself to a variety of applications in both academic and industrial settings. These cold dwelling bacteria are an abundant source of psychrophilic enzymes which have an interesting ability to retain higher catalytic activity at temperatures below 25 °C.[36][37] This is due to the highly malleable nature of these enzymes as this allows for better substrate - active site binding at colder temperatures.[36] This is important as enzymes that operate at lower temperatures not only make the industrial processes more efficient, but they also minimize the chance of side reactions occurring.[36][37] More of the substrate can directly be converted into the desired product all the while requiring less energy to do so. Psychrophilic enzymes can also aid with heat labile or volatile compounds, allowing reactions to occur without significant product loss.[36] Another unique application for these enzymes is the ability to be inhibited without the need of external reagents.[36] Usually to stop enzyme activity, chemical inhibitors are required which then require subsequent purification steps. With psychrophilic enzymes you can add slight heat to prevent any further reaction from occurring. Psychrophilic proteases derived from Polaribacter can be added to detergents allowing the washing of fabric at room temperature.[36]
An example of this is the enzyme carrageenase, which has been shown to have anti-tumor, antiviral, antioxidant and immunomodulatory activities. However, carrageenase isolated from bacteria has historically had low enzyme activity and poor stability.[38] Recently researchers have isolated and cloned the carrageenase gene from the Polaribacter sp. NJDZ03, which shows better thermostability, and the ability to be active at lower temperatures, making it a better choice for industrial uses.[38]
Exopolysaccharide
EPS is a secreted exopolysaccharide which protects the cells, stabilizes membranes, and serve and carbon stores.[39] Most EPS is similar but it is found that in extremophiles, the composition may be distinct.[39] Specifically in Polaribacter sp. SM1127, where the EPS has antioxidant activity and has shown to protect human fibroblast cells at lower temperatures.[39] Studies by Sun et al. were done to determine whether this can be utilized to protect and repair damage caused by frostbite.[39] It was found that Polaribacter derived EPS helps facilitate the dermal fibroblast cell movement to the site of injury. This not only promotes healing during frostbite injury but other cutaneous wounds as well/[39]
References
- Parte AC (2013). Parker CT, Garrity GM (eds.). "Polaribacter". NamesforLife. doi:10.1601/nm.8177.
- Bowman JP (2018-11-19). "Polaribacter". Bergey's Manual of Systematics of Archaea and Bacteria: 1–21. doi:10.1002/9781118960608.gbm00333.pub2. ISBN 9781118960608. S2CID 239787623.
- Park S, Park JM, Jung YT, Lee KC, Lee JS, Yoon JH (December 2014). "Polaribacter marinivivus sp. nov., a member of the family Flavobacteriaceae isolated from seawater". Antonie van Leeuwenhoek. 106 (6): 1139–1146. doi:10.1007/s10482-014-0283-4. PMID 25224356. S2CID 18062687.
- González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. (June 2008). "Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria)". Proceedings of the National Academy of Sciences of the United States of America. 105 (25): 8724–8729. doi:10.1073/pnas.0712027105. PMC 2438413. PMID 18552178.
- US EPA, OW (2013-09-05). "Climate Change and Harmful Algal Blooms". www.epa.gov. Retrieved 2022-04-06.
- Avcı B, Krüger K, Fuchs BM, Teeling H, Amann RI (June 2020). "Polysaccharide niche partitioning of distinct Polaribacter clades during North Sea spring algal blooms". The ISME Journal. 14 (6): 1369–1383. doi:10.1038/s41396-020-0601-y. PMC 7242417. PMID 32071394.
- "Genus: Polaribacter". www.bacterio.net. Retrieved 2022-02-16.
- Xu W, Chen XY, Wei XT, Lu DC, Du ZJ (March 2020). "Polaribacter aquimarinus sp. nov., isolated from the surface of a marine red alga". Antonie van Leeuwenhoek. 113 (3): 407–415. doi:10.1007/s10482-019-01350-z. PMID 31628626. S2CID 204757649.
- Malmstrom RR, Straza TR, Cottrell MT, Kirchman DL (2007-04-03). "Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean". Aquatic Microbial Ecology. 47: 45–55. doi:10.3354/ame047045. ISSN 0948-3055.
- Straza TR, Ducklow HW, Murray AE, Kirchman D (November 2010). "Abundance and single-cell activity of bacterial groups in Antarctic coastal waters". Limnology and Oceanography. 55 (6): 2526–2536. Bibcode:2010LimOc..55.2526S. doi:10.4319/lo.2010.55.6.2526. S2CID 86616866.
- Nikrad MP, Cottrell MT, Kirchman DL (April 2012). "Abundance and single-cell activity of heterotrophic bacterial groups in the western Arctic Ocean in summer and winter". Applied and Environmental Microbiology. 78 (7): 2402–2409. Bibcode:2012ApEnM..78.2402N. doi:10.1128/AEM.07130-11. PMC 3302604. PMID 22286998.
- Hahnke RL, Bennke CM, Fuchs BM, Mann AJ, Rhiel E, Teeling H, et al. (October 2015). "Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea". Environmental Microbiology. 17 (10): 3515–3526. doi:10.1111/1462-2920.12479. PMID 24725270.
- Cordone A, D'Errico G, Magliulo M, Bolinesi F, Selci M, Basili M, et al. (2022-01-27). "Bacterioplankton Diversity and Distribution in Relation to Phytoplankton Community Structure in the Ross Sea Surface Waters". Frontiers in Microbiology. 13: 722900. doi:10.3389/fmicb.2022.722900. PMC 8828583. PMID 35154048.
- Landa M, Blain S, Harmand J, Monchy S, Rapaport A, Obernosterer I (April 2018). "Major changes in the composition of a Southern Ocean bacterial community in response to diatom-derived dissolved organic matter". FEMS Microbiology Ecology. 94 (4). doi:10.1093/femsec/fiy034. PMID 29547927.
- Zeng YX, Yu Y, Qiao ZY, Jin HY, Li HR (February 2014). "Diversity of bacterioplankton in coastal seawaters of Fildes Peninsula, King George Island, Antarctica". Archives of Microbiology. 196 (2): 137–147. doi:10.1007/s00203-013-0950-2. PMID 24408126. S2CID 15257869.
- Comeau AM, Li WK, Tremblay JÉ, Carmack EC, Lovejoy C (2011-11-09). "Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum". PLOS ONE. 6 (11): e27492. Bibcode:2011PLoSO...627492C. doi:10.1371/journal.pone.0027492. PMC 3212577. PMID 22096583.
- Xing P, Hahnke RL, Unfried F, Markert S, Huang S, Barbeyron T, et al. (June 2015). "Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom". The ISME Journal. 9 (6): 1410–1422. doi:10.1038/ismej.2014.225. PMC 4438327. PMID 25478683.
- Gosink JJ, Woese CR, Staley JT (January 1998). "Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of 'Flectobacillus glomeratus' as Polaribacter glomeratus comb. nov". International Journal of Systematic Bacteriology. 48 (1): 223–235. doi:10.1099/00207713-48-1-223. PMID 9542092.
- Li H, Zhang XY, Liu C, Lin CY, Xu Z, Chen XL, et al. (March 2014). "Polaribacter huanghezhanensis sp. nov., isolated from Arctic fjord sediment, and emended description of the genus Polaribacter". International Journal of Systematic and Evolutionary Microbiology. 64 (Pt 3): 973–978. doi:10.1099/ijs.0.056788-0. PMID 24425815.
- Fukui Y, Abe M, Kobayashi M, Saito H, Oikawa H, Yano Y, Satomi M (May 2013). "Polaribacter porphyrae sp. nov., isolated from the red alga Porphyra yezoensis, and emended descriptions of the genus Polaribacter and two Polaribacter species". International Journal of Systematic and Evolutionary Microbiology. 63 (Pt 5): 1665–1672. doi:10.1099/ijs.0.041434-0. PMID 22904227.
- Nedashkovskaya OI, Kukhlevskiy AD, Zhukova NV (January 2013). "Polaribacter reichenbachii sp. nov.: a new marine bacterium associated with the green alga Ulva fenestrata". Current Microbiology. 66 (1): 16–21. doi:10.1007/s00284-012-0200-x. PMID 23053482. S2CID 18765393.
- Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, et al. (May 2013). "Key microbial drivers in Antarctic aquatic environments". FEMS Microbiology Reviews. 37 (3): 303–335. doi:10.1111/1574-6976.12007. PMID 23062173. S2CID 206922820.
- Luria CM, Amaral-Zettler LA, Ducklow HW, Rich JJ (2016). "Seasonal Succession of Free-Living Bacterial Communities in Coastal Waters of the Western Antarctic Peninsula". Frontiers in Microbiology. 7: 1731. doi:10.3389/fmicb.2016.01731. PMC 5093341. PMID 27857708.
- Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M, et al. (October 2012). "A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters". The ISME Journal. 6 (10): 1901–1915. doi:10.1038/ismej.2012.31. PMC 3446801. PMID 22534611.
- Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, et al. (January 2012). "Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean". Environmental Microbiology. 14 (1): 52–66. doi:10.1111/j.1462-2920.2011.02555.x. PMID 21895912.
- Yoon K, Song JY, Kwak MJ, Kwon SK, Kim JF (July 2017). "Genome characteristics of the proteorhodopsin-containing marine flavobacterium Polaribacter dokdonensis DSW-5". Journal of Microbiology. 55 (7): 561–567. doi:10.1007/s12275-017-6427-2. PMID 28432541. S2CID 8668865.
- Zhang Q, Fu L, Gui Y, Miao J, Li J (February 2022). "Complete genome sequence of Polaribacter sejongensis NJDZ03 exhibiting diverse macroalgal polysaccharide-degrading activity". Marine Genomics. 61: 100913. doi:10.1016/j.margen.2021.100913. PMID 35058032. S2CID 240140283.
- Kim E, Shin SK, Choi S, Yi H (January 2017). "Polaribacter vadi sp. nov., isolated from a marine gastropod". International Journal of Systematic and Evolutionary Microbiology. 67 (1): 144–147. doi:10.1099/ijsem.0.001591. PMID 27902220.
- Hyun DW, Shin NR, Kim MS, Kim PS, Jung MJ, Kim JY, et al. (May 2014). "Polaribacter atrinae sp. nov., isolated from the intestine of a comb pen shell, Atrina pectinata". International Journal of Systematic and Evolutionary Microbiology. 64 (Pt 5): 1654–1661. doi:10.1099/ijs.0.060889-0. PMID 24510977.
- Parker, Charles Thomas; Garrity, George M. (2013). Parker, Charles Thomas; Garrity, George M (eds.). "Polaribacter". doi:10.1601/nm.8177.
{{cite journal}}: Cite journal requires|journal=(help) - Park S, Choi SJ, Choi J, Yoon JH (November 2017). "Paraglaciecola aestuariivivens sp. nov., isolated from a tidal flat". International Journal of Systematic and Evolutionary Microbiology. 67 (11): 4754–4759. doi:10.1099/ijsem.0.002370. PMID 28984552.
- Choo S, Borchert E, Wiese J, Saha M, Künzel S, Weinberger F, Hentschel U (July 2020). "Polaribacter septentrionalilitoris sp. nov., isolated from the biofilm of a stone from the North Sea". International Journal of Systematic and Evolutionary Microbiology. 70 (7): 4305–4314. doi:10.1099/ijsem.0.004290. PMID 32579104. S2CID 220049277.
- Kang I, Jang H, Cho JC (December 2015). "Complete genome sequences of bacteriophages P12002L and P12002S, two lytic phages that infect a marine Polaribacter strain". Standards in Genomic Sciences. 10 (1): 82. doi:10.1186/s40793-015-0076-z. PMC 4615864. PMID 26500718.
- Bartlau N, Wichels A, Krohne G, Adriaenssens EM, Heins A, Fuchs BM, Amann R, Moraru C (2021-05-20). "Highly diverse flavobacterial phages as mortality factor during North Sea spring blooms". bioRxiv. doi:10.1101/2021.05.20.444936. S2CID 235097164.
- Urbanek AK, Rymowicz W, Mirończuk AM (September 2018). "Degradation of plastics and plastic-degrading bacteria in cold marine habitats". Applied Microbiology and Biotechnology. 102 (18): 7669–7678. doi:10.1007/s00253-018-9195-y. PMC 6132502. PMID 29992436.
- Annapure US, Pratisha N (January 2022). "Chapter 14 - Psychrozymes: A novel and promising resource for industrial applications". In Kuddus M (ed.). Microbial Extremozymes. Academic Press. pp. 185–195. doi:10.1016/B978-0-12-822945-3.00018-X. ISBN 978-0-12-822945-3. Retrieved 2022-04-08.
- Feller G, Gerday C (December 2003). "Psychrophilic enzymes: hot topics in cold adaptation". Nature Reviews. Microbiology. 1 (3): 200–208. doi:10.1038/nrmicro773. PMID 15035024. S2CID 6441046.
- Gui Y, Gu X, Fu L, Zhang Q, Zhang P, Li J (2021). "Expression and Characterization of a Thermostable Carrageenase From an Antarctic Polaribacter sp. NJDZ03 Strain". Frontiers in Microbiology. 12: 631039. doi:10.3389/fmicb.2021.631039. PMC 7994522. PMID 33776960.
- Sun ML, Zhao F, Chen XL, Zhang XY, Zhang YZ, Song XY, et al. (January 2020). "Promotion of Wound Healing and Prevention of Frostbite Injury in Rat Skin by Exopolysaccharide from the Arctic Marine Bacterium Polaribacter sp. SM1127". Marine Drugs. 18 (1): 48. doi:10.3390/md18010048. PMC 7024241. PMID 31940773.
Further reading
- Hyun DW, Shin NR, Kim MS, Kim PS, Jung MJ, Kim JY, et al. (May 2014). "Polaribacter atrinae sp. nov., isolated from the intestine of a comb pen shell, Atrina pectinata". International Journal of Systematic and Evolutionary Microbiology. 64 (Pt 5): 1654–1661. doi:10.1099/ijs.0.060889-0. PMID 24510977.
- Nedashkovskaya OI, Kim SB, Lysenko AM, Kalinovskaya NI, Mikhailov VV, Kim IS, Bae KS (December 2005). "Polaribacter butkevichii sp. nov., a novel marine mesophilic bacterium of the family Flavobacteriaceae". Current Microbiology. 51 (6): 408–412. doi:10.1007/s00284-005-0105-z. PMID 16235024. S2CID 27074806.
- Gosink JJ, Woese CR, Staley JT (January 1998). "Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of 'Flectobacillus glomeratus' as Polaribacter glomeratus comb. nov". International Journal of Systematic Bacteriology. 48 (1): 223–235. doi:10.1099/00207713-48-1-223. PMID 9542092.
- Kim E, Shin SK, Choi S, Yi H (January 2017). "Polaribacter vadi sp. nov., isolated from a marine gastropod". International Journal of Systematic and Evolutionary Microbiology. 67 (1): 144–147. doi:10.1099/ijsem.0.001591. PMID 27902220.
- Xu W, Chen XY, Wei XT, Lu DC, Du ZJ (March 2020). "Polaribacter aquimarinus sp. nov., isolated from the surface of a marine red alga". Antonie van Leeuwenhoek. 113 (3): 407–415. doi:10.1007/s10482-019-01350-z. PMID 31628626. S2CID 204757649.
- Kim YO, Park IS, Park S, Nam BH, Park JM, Kim DG, Yoon JH (December 2016). "Polaribacter haliotis sp. nov., isolated from the gut of abalone Haliotis discus hannai". International Journal of Systematic and Evolutionary Microbiology. 66 (12): 5562–5567. doi:10.1099/ijsem.0.001557. PMID 27902190.