Polyphyly
A polyphyletic group is an assemblage that includes organisms with mixed evolutionary origin but does not include their most recent common ancestor.[1] The term is often applied to groups that share similar features known as homoplasies, which are explained as a result of convergent evolution. The arrangement of the members of a polyphyletic group is called a polyphyly /ˈpɒlɪˌfaɪli/.[2] It is contrasted with monophyly and paraphyly.
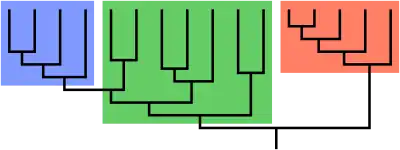
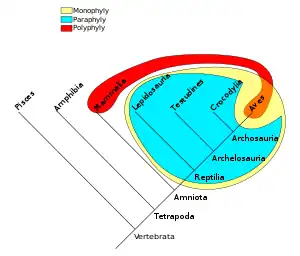
For example, the biological characteristic of warm-bloodedness evolved separately in the ancestors of mammals and the ancestors of birds; "warm-blooded animals" is therefore a polyphyletic grouping.[3] Other examples of polyphyletic groups are algae, C4 photosynthetic plants,[4] and edentates.[5]
Many taxonomists aim to avoid homoplasies in grouping taxa together, with a goal to identify and eliminate groups that are found to be polyphyletic. This is often the stimulus for major revisions of the classification schemes. Researchers concerned more with ecology than with systematics may take polyphyletic groups as legitimate subject matter; the similarities in activity within the fungus group Alternaria, for example, can lead researchers to regard the group as a valid genus while acknowledging its polyphyly.[6] In recent research, the concepts of monophyly, paraphyly, and polyphyly have been used in deducing key genes for barcoding of diverse groups of species.[7]
Etymology
The term polyphyly, or polyphyletic, derives from the two Ancient Greek words, πολύς (polús), meaning "many, a lot of", and φῦλον (phûlon), meaning "genus, species",[8][9] and refers to the fact that a polyphyletic group includes organisms (e.g., genera, species) arising from multiple ancestral sources.
Conversely, the term monophyly, or monophyletic, builds on the ancient Greek prefix μόνος (mónos), meaning "alone, only, unique",[8][9] and refers to the fact that a monophyletic group includes organisms consisting of all the descendants of a unique common ancestor.
By comparison, the term paraphyly, or paraphyletic, uses the ancient Greek prefix παρά (pará), meaning "beside, near",[8][9] and refers to the situation in which one or several monophyletic subgroups are left apart from all other descendants of a unique common ancestor.
Avoidance
In many schools of taxonomy, the recognition of polyphyletic groups in a classification is discouraged. Monophyletic groups (that is, clades) are considered by these schools of thought to be the only valid groupings of organisms because they are diagnosed ("defined", in common parlance) on the basis of synapomorphies, while paraphyletic or polyphyletic groups are not. From the perspective of ancestry, clades are simple to define in purely phylogenetic terms without reference to clades previously introduced: a node-based clade definition, for example, could be "All descendants of the last common ancestor of species X and Y". On the other hand, polyphyletic groups can be delimited as a conjunction of several clades, for example "the flying vertebrates consist of the bat, bird, and pterosaur clades".
From a practical perspective, grouping species monophyletically facilitates prediction far more than does polyphyletic grouping. For example, classifying a newly discovered grass in the monophyletic family Poaceae, the true grasses, immediately results in numerous predictions about its structure and its developmental and reproductive characteristics, that are synapomorphies of this family. In contrast, Linnaeus' assignment of plants with two stamens to the polyphyletic class Diandria, while practical for identification, turns out to be useless for prediction, since the presence of exactly two stamens has developed convergently in many groups.[10]
Polyphyletic species
Species have a special status in systematics as being an observable feature of nature itself and as the basic unit of classification.[11] It is usually implicitly assumed that species are monophyletic (or at least paraphyletic). However, hybrid speciation arguably leads to polyphyletic species.[12] Hybrid species are a common phenomenon in nature, particularly in plants where polyploidy allows for rapid speciation.[13] Some cladist authors do not consider species to possess the property of "-phyly", which they assert applies only to groups of species.[14][15]
See also
References
- Urry, Lisa A. (2016). Campbell Biology (11th ed.). Pearson. p. 558. ISBN 978-0-134-09341-3.
- "polyphyly". Oxford English Dictionary (Online ed.). Oxford University Press. Retrieved 28 December 2021. (Subscription or participating institution membership required.). [Source for pronunciation.]
- Archibald, J. David (2014-07-15). Aristotle's Ladder, Darwin's Tree: The Evolution of Visual Metaphors for Biological Order. Columbia University Press. ISBN 9780231164122.
- Sage, Rowan F. (2004-02-01). "The evolution of C4 photosynthesis". New Phytologist. 161 (2): 341–370. doi:10.1111/j.1469-8137.2004.00974.x. ISSN 1469-8137. PMID 33873498.
- Delsuc, Frédéric; Douzery, Emmanuel J. P. (2008). "Recent advances and future prospects in xenarthran molecular phylogenetics". In Vizcaíno, Sergio F.; Loughry, W. J. (eds.). The biology of the Xenarthra. Gainesville: University Press of Florida. pp. 11–23. ISBN 9780813031651. OCLC 741613153.
- Aschehoug, Erik T.; Metlen, Kerry L.; Callaway, Ragan M.; Newcombe, George (2012). "Fungal endophytes directly increase the competitive effects of an invasive forb". Ecology. 93 (1): 3–8. doi:10.1890/11-1347.1. PMID 22486080.
- Parhi J., Tripathy P.S., Priyadarshi, H., Mandal S.C., Pandey P.K. (2019). "Diagnosis of mitogenome for robust phylogeny: A case of Cypriniformes fish group". Gene. 713: 143967. doi:10.1016/j.gene.2019.143967. PMID 31279710. S2CID 195828782.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Bailly, Anatole (1981-01-01). Abrégé du dictionnaire grec français. Paris: Hachette. ISBN 978-2010035289. OCLC 461974285.
- Bailly, Anatole. "Greek-french dictionary online". www.tabularium.be. Retrieved March 2, 2018.
- Stace, Clive A. (2010). "Classification by molecules: What's in it for field botanists?" (PDF). Watsonia. 28: 103–122. Archived from the original (PDF) on October 15, 2012. Retrieved July 31, 2013.
- Queiroz, Kevin; Donoghue, Michael J. (December 1988). "Phylogenetic Systematics and the Species Problem". Cladistics. 4 (4): 317–338. doi:10.1111/j.1096-0031.1988.tb00518.x. PMID 34949064. S2CID 40799805.
- Hörandl, E.; Stuessy, T.F. (2010). "Paraphyletic groups as natural units of biological classification". Taxon. 59 (6): 1641–1653. doi:10.1002/tax.596001.
- Linder, C.R.; Risenberg, L.H. (22 June 2004). "Reconstructing patterns of reticulate evolution in plants". American Journal of Botany. 91 (10): 1700–1708. doi:10.3732/ajb.91.10.1700. PMC 2493047. PMID 18677414.
- Nixon, Kevin C., and Quentin D. Wheeler. "An amplification of the phylogenetic species concept." Cladistics 6, no. 3 (1990): 211–223.
- Brower, Andrew V.Z., and Randall T. Schuh. 2021. "Biological Systematics: Principles and Applications" (3rd edn.). Cornell University Press, Ithaca, NY.
Bibliography
- Tudge, Colin (2000). The Variety of Life. Oxford University Press. ISBN 0-19-860426-2.
- "Evolution - A-Z - Polyphyletic group". www.blackwellpublishing.com. Retrieved 2018-02-24.
External links
- Funk, D. J., and Omland, K. E. (2003). "Species-level paraphyly and polyphyly: Frequency, cause and consequences, with insights from animal mitochondrial DNA" Annu. Rev. Ecol. Evol. Syst. 34: 397–423. at ftp://137.110.142.4/users/bhhanser/Subspecies%20general%20literature/FunkEtal2003AnnuRevEcolEvolV34pp397-423.pdf%5B%5D
