Prasinovirus
Prasinovirus is a genus of large double-stranded DNA viruses, in the family Phycodnaviridae that infect phytoplankton in the Prasinophyceae. There are three groups in this genus,[1][2] including Micromonas pusilla virus SP1, which infects the cosmopolitan photosynthetic flagellate Micromonas pusilla.[3]
| Prasinovirus | |
|---|---|
 | |
| Negative stained image of MpV-SP1 | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Varidnaviria |
| Kingdom: | Bamfordvirae |
| Phylum: | Nucleocytoviricota |
| Class: | Megaviricetes |
| Order: | Algavirales |
| Family: | Phycodnaviridae |
| Genus: | Prasinovirus |
There is a large group of genetically diverse but related viruses that show considerable evidence of lateral gene transfer.[4][5]
Taxonomy

The genus contains the following species:[2]
- Micromonas pusilla virus MpVs
- Ostreococcus tauri virus OtVs
- Ostreococcus lucimarinus virus OlVs
- Bathycoccus prasino virus BpVs
- Bathycoccus sp. clade II virus BIIVs
Structure
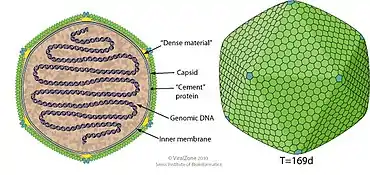
Viruses in Prasinovirus are enveloped, with icosahedral and round geometries, and T=169 symmetry. The diameter is around 104-118 nm.[1]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Prasinovirus | Icosahedral | T=169 | Enveloped | Linear | Monopartite |
Life cycle
Viral replication is nucleo-cytoplasmic. Replication follows the DNA strand displacement model. DNA templated transcription is the method of transcription. The virus exits the host cell by lysis via lytic phospholipids. Alga serve as the natural host. Transmission routes are passive diffusion.[1]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Prasinovirus | Alga | None | Cell receptor endocytosis | Lysis | Nucleus | Cytoplasm | Passive diffusion |
References
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 22 May 2021.
- Cottrell, Matthew T.; Suttle, Curtis A. (1991). "Wide-spread occurrence and clonal variation in viruses which cause lysis of a cosmopolitan, eukaryotic marine phytoplankter, "Micromonas pusilla"". Marine Ecology Progress Series. 78: 1–9. Bibcode:1991MEPS...78....1C. doi:10.3354/meps078001. ISSN 1616-1599.
- Bellec, Laure; Grimsley, Nigel; Derelle, Evelyn; Moreau, Herve; Desdevises, Yves (2010). "Abundance, spatial distribution and genetic diversity of Ostreococcus tauri viruses in two different environments". Environmental Microbiology Reports. 2 (2): 313–321. doi:10.1111/j.1758-2229.2010.00138.x. PMID 23766083.
- Finke, Jan F; Winget, Danielle M; Chan, Amy M; Suttle, Curtis A (2017). "Variation in the genetic repertoire of viruses Infecting Micromonas pusilla reflects horizontal gene transfer and links to their environmental distribution". Viruses. 9 (5): 116. doi:10.3390/v9050116. PMC 5454428. PMID 28534829.