Rhodovulum sulfidophilum
Rhodovulum sulfidophilum is a gram-negative purple nonsulfur bacteria.[1] The cells are rod-shaped, and range in size from 0.6 to 0.9 μm wide and 0.9 to 2.0 μm long, and have a polar flagella. These cells reproduce asexually by binary fission. This bacterium can grow anaerobically (photoautotrophic or photoheterotrophic) when light is present, or aerobically (chemoheterotrophic) under dark conditions.[2][3] It contains the photosynthetic pigments bacteriochlorophyll a and of carotenoids.[1]
| Rhodovulum sulfidophilum | |
|---|---|
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | R. sulfidophilum |
| Binomial name | |
| Rhodovulum sulfidophilum Hiraishi and Ueda, 1994 | |
There is interest in R. sulfidophilum for its applications in producing artificial spider silk [4] and biocompounds,[5] waste remediation[6] and for its use in aquaculture.[7]
Taxonomy
Rhodovulum sulfidophilum was first reported by Hansen and Veldkamp in 1973, and given the name Rhodopseudomonas sulfidophila.[1] The bacterium was later reclassified to the Rhodovulum genus in 1994 by Hiraishi and Ueda, as all marine Rhodobacter and Rhodopseudomonas species were transferred to the genus Rhodovulum due to marine Rhodobacter species being able to be differentiated phenotypically from the freshwater species. The Rhodovulum genus have been described with additional properties of G+C content of the DNA that ranges from 66.3 to 66.6 mol% in type strain DSM 1374.[8]
Genome
A genome draft for R. sulfidophilum DSM 1374 was published in 2013, and was predicted to have a chromosome with 3983 genes that code for protein, and two circular plasmids with 93 and 84 genes respectively.[9] The first complete genome was published for strain DSM 2351, revealing a circular chromosome and three circular plasmids of differing DNA base-pair size.[10] Additionally, draft genomes of the AB14, AB26 and AB30 strains were published, and when compared showed diversity between the different strains of R. sulfidophilum.[11] These draft genomes also revealed that they contained genes confirmed to be involved in cellular processes, such as carbon dioxide fixation and sulfur oxidation.[11] Strain AB26 was discovered to have a unique 100 kb plasmid sequence homologous to plasmid 3 that contain genes related to metal metabolism and transport (e.g. manganese, nickel and zinc transport proteins).[11] A later study that analyzed isolates of R. sulfidophilum found that AB26 reserved around 16% of its entire 4,380,746 base pair genome to transport and also had 20 response regulators, and 22 histidine kinases.[12] Another study that genetically characterized R. sulfidophilum discovered that the bacterium contained one copy of pucA and pucB (α- and β- genes) each within the puc operon.[13] In addition, it was found that there were no Integration Host Factor (IHF) and Fumerate and Nitrate Reductase (FNR) regulation protein binding sites in this bacterial species like with Rhodobacter sphaeroides.[13] It was speculated that the lack of binding sites for these proteins are indicative of why the puc operon is expressed when oxygen is present.[13] It was also discovered that while the mRNA sequences between R. sphaeroides and R. sulfidophilum were homologous, a regulatory region exhibited an exchange of TGT to CGT in R. sulfidophilum.[13] Studying the genome of R. sulfidophilum is of particular interest for better understanding relevant metabolic pathways.
When analysing evolution and horizontal gene transfer of Proteobacteria photosynthetic gene clusters, it was shown that R. sulfidophilum shares regions of genetic similarity to a group of other species that range from Roseibacterium elongatum to Dineoroseobacter shibae.[14]
Metabolism
Rhodovulum sulfidophilum is metabolically versatile and can adapt to various environmental conditions by using various metabolic pathways under anaerobic conditions.[1] Primary sources of energy for R. sulfidophilum consist of light, organic compounds and inorganic sulfur compounds.[1][11]
When provided with a light source and nitrogen, using either malate as a carbon source or sulfide as an electron source, it can undergo photosynthesis to produce ATP.[15] Rhodovulum sulfidophilum is capable of oxidizing sulfide or thiosulfate to yield sulfates in the ecosystem without accumulating intermediates and have an unusual tri-heme cytochrome subunit bound to the reaction center, therefore allowing for photolithoautotrophic growth.[9] Oxidation of thiosulfate is done by the SoxAX protein, which is a heterodimeric c-type cytochrome.[1][16] Additionally, unlike other nonsulfur purple bacteria, R. sulfidophilum is able to synthesize its peripheral antenna complex under dark aerobic conditions.[13] Rhodovulum sulfidophilum can utilize a wide variety of organic compunds, and grows both photoorganotrophically and heterotrophically.[13]
Multiple strains of R. sulfidophilum have also been shown to be capable of photoferrotrophy, a process that fixes inorganic carbon to organic material using light and Fe(II) as the electron donor.[17][18] Rhodovulum sulfidophilum DSM 2351 is found to produce and excrete high levels of nucleic acids, which causes cell aggregates to form.[19] Additionally, photoferrotrophic organisms like R. sulfidophilum are capable of using specific conductive minerals as electron donors through phototrophic extracellular electron uptake (pEEu).[12]
Habitat and ecology
Habitat
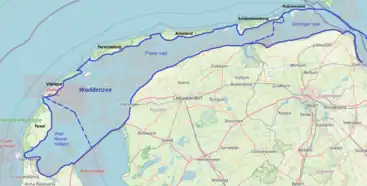
Rhodovulum sulfidophilum has been found in a variety of aquatic environments, including freshwater pelagic and benthic habitats. Rhodovulum sulfidophilum was originally isolated from the mud of the marine floor of the intertidal flats of the Dutch Waddenzee, north of the province of Groningen, Netherlands.[1]
The particular geographic region or climate were Rhodovulum sulfidophilum can be located is dependent on growth dependent factors such as salinity, temperature, and other environmental conditions. One study discovered this species of bacteria to be abundant in the sediment surface layer of a freshwater pond while another experiment isolated it from the anaerobic sludge of Hongdao shrimp pond, located in Qingdao, China.[20] Another study isolated Rhodovulum sulfidophilum from a microbial mat within a brackish estuary in Massachusetts, USA.[11] Additional studies also discovered R. sulfidophilum in seashore sediment samples from Osaka Bay and tidal water samples from a beach at Visakhapatnam in coastal areas of Andhra Pradesh, India.[21][22]
Distribution
Due to its metabolic flexibility, Rhodovulum sulfidophilum exhibits a relatively wide distribution and is found in a variety of aquatic habitats, especially anaerobic environments with high sulfide concentrations. The main environmental factor governing the growth of purple bacteria species such as Rhodovulum sulfidophilum. is the presence and concentration of sulfide.[23] While other Purple Non-Sulfur Bacteria (PNSB) species are incapable of surviving in sulfur-rich areas, these conditions are ideal for R.sulfidophilum as it utilizes sulfide as the donor for electrons when conducting photolithotrophic metabolic processes. Another factor contributing to the proliferation of this bacterium is organic matter concentrations in water, since it can also undergo photoorganotrophic growth.[23] Therefore in wastewater regions with a large chemical oxygen demand, Rhodovulum sulfidophilum is still capable of growth.[24] Based on these conditions, R. sulfidophilum is typically found in marine environments with sulfidic conditions, such as marine sediments, seawater pools, mud flats, tidal areas, and sulfur-rich coastal waters. [1][20][22]
R. sulfidophilum has been discovered to optimally grow in areas that have a temperature range of 30-35 °C and at a pH of around 7.0.[25] Anaerobic conditions are necessary for Rhodovulum sulfidophilum's growth and to use a variety of organic compounds such as lactic, butyric, and acetic acid as carbon and energy sources in low oxygen conditions with available light.[25] It can also use bacteriochlorophyll a and sulfur oxidation to perform photosynthesis as an adaptation to anaerobic climates with high sulfide concentrations.
Possible role in climate change
Much of the greenhouse gas produced by anthropogenic activities is stored within the ocean, which is the largest existing reservoir of carbon dioxide on earth.[26] Since metabolically flexible microbes such as Rhodovulum sulfidophilum are capable of fixing carbon using multiple different pathways (e.g. ferrous iron oxidation, pEEu), researchers have begun conducting studies concerning the extent by which photoferrotrophic freshwater microbes contribute to mitigating fluctuations in marine carbon concentrations via carbon sequestration.[18] Considering that marine sediments account for about fifty percent of global primary production, it may be worthwhile looking into how photoferrotrophic organisms such as R. sulfidophilum could contribute to reducing the amount of excess carbon within the ocean.[27][18]
Viral infection
Rhodovulum sulfidophilum contains a prophage.[28] It has been used as a model organism for lysogenic bacteria in studies regarding the role of viruses in marine biogeochemical cycles. Viruses that infect Rhodovulum sulfidophilum include bacteriophage members of the Siphoviridae family, such as Rhodovulum phage RS1 (GenBank: JF974307.1) that has a tail length of 100 nm and a capsid diameter of 46 nm.[29] Rhodovulum sulfidophilum strain P122 A, which has been cultured from deep-subseafloor sediments, has two known phages with 36kb genomes and a head/tail of 46/100 mm.[28]
Applications
Production of spider silk
_artificial_spidroin.webp.png.webp)
Spider silk is a strong biodegradable material well-suited for biomedical uses. However, due to the cannibalistic nature of spiders and low silk potential in spider glands, silk yields are low. Genetic approaches to increase silk production yields have been attempted.[4] The required spider genes have been successfully recombined in heterotrophic organisms ranging from bacteria (E.coli) to animals (mice). However, these microbial cell factories are unsustainable and costly due to the high metabolic demands for organic materials.[5]
In 2020, a research team in Japan identified R. sulfidophilum as a sustainable and low cost silk producing microbial cell factory. Genetically modified R. sulfidophilum can produce the hydrophobic repetitive sequence of major ampullate spidroin (MaSp), the major protein in spider silk. After constructing a plasmid containing the necessary genetic information for the MaSp1 gene, researchers conjugated the plasmid into R. sulfidophilum and observed gene expression and spider silk fiber formation in heterotrophic and autotrophic growth conditions.[5] Notably, as a photoautotroph, the marine bacteria uses low-cost, abundant and renewable resources to make the silk: CO2 as a carbon source, light as an energy source and N2 as a nitrogen source. While more work is necessary, R. sulfidophilum proves to be a sustainable, cost-effective photosynthetic microbial cell factory for artificial silk production.[30]
Production of biodegradable plastic
Given that petroleum-derived plastics are expected to persist within the environment when discarded and therefore pose a threat as a widespread pollutant, continuous research is being conducted on the production bioplastic materials as a sustainable alternative.[31] Biopolyesters, such as polyhydroxyalkanoates (PHAs), are sustainable alternatives to petroleum-based plastics as they are biodegradable, biocompatible, and can also be produced biologically.[32][33] PHAs are a metabolic result of excess carbon.[34] This production is normally costly due to high carbon demands, however R. sulfidophilum proves to be an effective alternative as it generates its own carbon source through photosynthesis.
Rhodovulum sulfidophilum has the ability to synthesize polyhydroxyalkanoate (PHA) within bacterial cells.[35] This bacterium does so via photo-fermentation which allows it to transform specific organic acids (OAs) such as succinate, lactate, and malate as single carbon sources into the following well known polymers of the PHA family, P3HB (poly-3-hydroxybutyrate) and P3HB-co-3 HV (3-hydroxybutyrateco-3-hydroxyvalerate).[32] Additionally, a PHA synthase (PhaC) gene is present in the R. sulfidophilum genome. Anoxygenic photosynthetic bacteria have been shown to produce higher amounts of PHA compared to oxygenic phototrophs, such as plants and cyanobacteria.[35]
Production of H2
As a photoheterotrophic bacteria, R. sulfidophilum can anaerobically produce H2 from low cost organic materials in presence of light. This is reaction is catalyzed by nitrogenase which is inhibited by oxygen, ammonia and a high N:C ratio. H2 is an emerging, but controversial form of renewable energy.[20] This process can also be used in the treatment of waste from agriculture, forestry and food processing.[25] Additionally, the hydrogen production rate of R. sulfidophilum be maximized by rapid degradation and use of endogenous substrate poly(3-hydroxybutyrate) (PHB) which is one of the most preferable substrates for hydrogen production in this strain. It can be achieved in high-density suspension under external-substrate-depleted conditions after aerobic cultivation in the presence of an excess amount of acetate. [36]
Waste bioremediation
As an anoxygenic sulfate reducing phototroph, R. sulfidophilum is used for bioremediation as it can grow in polluted environments, such as industrial fish processing wastewater.[24] Organic contamination often depletes oxygen and other terminal electron acceptors (TEA) in water and soil environments. Because of its low solubility, it is difficult to add sufficient oxygen to serve as a TEA in contaminated waters. The use of microbes to help metabolise the pollutant is an effective solution. The metabolism of the nutrients can provide substances that confer useful advantages to other organisms in the environment, which ultimately increases productivity.[6]
Antiviral properties
Aptamers are used as biosensors to detect diseases and act as therapeutics, such as in fisheries.[37] Rodovulum sulfidophilum can produce RNA antiviral aptamers against fish infecting viruses, such as hemorrhagic septicemia virus (VHSV) and hirame rhabdovirus.[38][39] These aptamers have been shown to reduce Japanese flounder mortality from 90% to 10% in 10 days post-infection. Rhodovulum sulfidophilum's aptamers have the potential to improve biosecurity in aquaculture and fisheries.[39] Additionally, this can be applied to recombinant RNA production technology using R. sulfidophilum.[3]
Probiotics
Purple non-sulphur bacteria, such as R. sulfidophilum, have been used in aquaculture as probiotics, which are microorganisms that have health benefits.[40] Kuruma shrimp (Marsupenaeus japonicus) is an important species in the global aquaculture industry, most notably being cultured in Japan and China. Probiotics have been used as an eco-friendly, low-cost approach to boost shrimp survival by upregulating immune system genes, such as antimicrobial peptides, and moulting-related genes, including cuticle and calcification proteins. Rhodovulum sulfidophilum can be effective as a probiotic at a low concentration (10^3 cfu/mL) in rearing waters. After the addition of R. sulfidophilum various features of shrimp growth are improved: body weight (by 1.76-fold), survival rate (by 8.3%), and the feed conversion ratio (by 10%).[7] Additionally, R. sulfidophilum has been shown to enhance the larval survival of marble goby (Oxyeleotris marmorat) and the performance of milkfish (Chanos chanos). Use of the beneficial bacteria, R. sulfidophilum, confers ammonia reduction and total phosphorus stabilization. This can help reduce the proliferation of cyanobacteria and algae blooms and eutrophication and promote the growth of other helpful bacteria, such as Acidobacteria.[41] Ultimately, R. sulfidophilum is used a probiotic to increase aquaculture efficiency and improve management.
References
- Hansen TA, Veldkamp H (1973). "Rhodopseudomonas sulfidophila, nov. spec., a new species of the purple nonsulfur bacteria". Archiv für Mikrobiologie. 92 (1): 45–58. doi:10.1007/BF00409510. PMID 4725822. S2CID 25160261.
- Imhoff JF, Kramer M, Trüper HG (1983). "Sulfate assimilation in Rhodopseudomonas sulfidophila". Archives of Microbiology. 136 (2): 96–101. doi:10.1007/BF00404780. ISSN 0302-8933. S2CID 9491589.
- Kikuchi Y, Umekage S (February 2018). "Extracellular nucleic acids of the marine bacterium Rhodovulum sulfidophilum and recombinant RNA production technology using bacteria". FEMS Microbiology Letters. 365 (3). doi:10.1093/femsle/fnx268. PMID 29228187.
- Ramezaniaghdam M, Nahdi ND, Reski R (2022-03-08). "Recombinant Spider Silk: Promises and Bottlenecks". Frontiers in Bioengineering and Biotechnology. 10: 835637. doi:10.3389/fbioe.2022.835637. PMC 8957953. PMID 35350182.
- Foong CP, Higuchi-Takeuchi M, Malay AD, Oktaviani NA, Thagun C, Numata K (July 2020). "A marine photosynthetic microbial cell factory as a platform for spider silk production". Communications Biology. 3 (1): 357. doi:10.1038/s42003-020-1099-6. PMC 7343832. PMID 32641733.
- Kobayashi M, Kobayashi M (2004). "Waste Remediation and Treatment Using Anoxygenic Phototrophic Bacteria". In Blankenship RE, Madigan MT, Bauer CE (eds.). Anoxygenic Photosynthetic Bacteria. Advances in Photosynthesis and Respiration. Vol. 2. Dordrecht: Kluwer Academic Publishers. pp. 1269–1282. doi:10.1007/0-306-47954-0_62. ISBN 978-0-7923-3681-5.
- Koga A, Goto M, Hayashi S, Yamamoto S, Miyasaka H (January 2022). "Probiotic Effects of a Marine Purple Non-Sulfur Bacterium, Rhodovulum sulfidophilum KKMI01, on Kuruma Shrimp (Marsupenaeus japonicus)". Microorganisms. 10 (2): 244. doi:10.3390/microorganisms10020244. PMC 8876596. PMID 35208699.
- Hiraishi A, Ueda Y (January 1994). "Intrageneric Structure of the Genus Rhodobacter: Transfer of Rhodobacter sulfidophilus and Related Marine Species to the Genus Rhodovulum gen. nov". International Journal of Systematic Bacteriology. 44 (1): 15–23. doi:10.1099/00207713-44-1-15. ISSN 0020-7713.
- Masuda S, Hori K, Maruyama F, Ren S, Sugimoto S, Yamamoto N, et al. (August 2013). "Whole-Genome Sequence of the Purple Photosynthetic Bacterium Rhodovulum sulfidophilum Strain W4". Genome Announcements. 1 (4): e00577-13. doi:10.1128/genomeA.00577-13. PMC 3738892. PMID 23929476.
- Nagao N, Hirose Y, Misawa N, Ohtsubo Y, Umekage S, Kikuchi Y (April 2015). "Complete Genome Sequence of Rhodovulum sulfidophilum DSM 2351, an Extracellular Nucleic Acid-Producing Bacterium". Genome Announcements. 3 (2): e00388-15. doi:10.1128/genomeA.00388-15. PMC 4417702. PMID 25931606. S2CID 36467883.
- Guzman MS, McGinley B, Santiago-Merced N, Gupta D, Bose A (March 2017). "Draft Genome Sequences of Three Closely Related Isolates of the Purple Nonsulfur Bacterium Rhodovulum sulfidophilum". Genome Announcements. 5 (11): e00029–17. doi:10.1128/genomeA.00029-17. PMC 5356053. PMID 28302776.
- Davenport EJ, Bose A (June 2022). "Taxonomic Re-Evaluation and Genomic Comparison of Novel Extracellular Electron Uptake-Capable Rhodovulum visakhapatnamense and Rhodovulum sulfidophilum Isolates". Microorganisms. 10 (6): 1235. doi:10.3390/microorganisms10061235. PMC 9230146. PMID 35744753.
- Hagemann GE, Katsiou E, Forkl H, Steindorf AC, Tadros MH (April 1997). "Gene cloning and regulation of gene expression of the puc operon from Rhodovulum sulfidophilum". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1351 (3): 341–358. doi:10.1016/s0167-4781(96)00228-x. PMID 9130598.
- Brinkmann H, Göker M, Koblížek M, Wagner-Döbler I, Petersen J (August 2018). "Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae". The ISME Journal. 12 (8): 1994–2010. doi:10.1038/s41396-018-0150-9. PMC 6052148. PMID 29795276.
- Kelley BC, Jouanneau Y, Vignais PM (August 1979). "Nitrogenase activity in Rhodopseudomonas sulfidophila". Archives of Microbiology. 122 (2): 145–152. doi:10.1007/bf00411353. ISSN 0302-8933. S2CID 32515373.
- Bamford VA, Bruno S, Rasmussen T, Appia-Ayme C, Cheesman MR, Berks BC, Hemmings AM (November 2002). "Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme". The EMBO Journal. Oxford University Press. 21 (21): 5599–610. doi:10.1093/emboj/cdf566. OCLC 678225926. PMC 131063. PMID 12411478.
- Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B, Schink B (April 1993). "Ferrous iron oxidation by anoxygenic phototrophic bacteria". Nature. 362 (6423): 834–836. doi:10.1038/362834a0. ISSN 0028-0836. S2CID 4261907.
- Gupta D, Guzman MS, Rengasamy K, Stoica A, Singh R, Ranaivoarisoa TO, et al. (November 2021). "Photoferrotrophy and phototrophic extracellular electron uptake is common in the marine anoxygenic phototroph Rhodovulum sulfidophilum". The ISME Journal. 15 (11): 3384–3398. doi:10.1038/s41396-021-01015-8. PMC 8528915. PMID 34054125.
- Suzuki, Hiromichi; Daimon, Masahide; Awano, Tomoyuki; Umekage, So; Tanaka, Terumichi; Kikuchi, Yo (2009-08-01). "Characterization of extracellular DNA production and flocculation of the marine photosynthetic bacterium Rhodovulum sulfidophilum". Applied Microbiology and Biotechnology. 84 (2): 349–356. doi:10.1007/s00253-009-2031-7. ISSN 1432-0614. PMID 19452150. S2CID 13006036.
- Cai J, Wang G (October 2012). "Hydrogen production by a marine photosynthetic bacterium, Rhodovulum sulfidophilum P5, isolated from a shrimp pond". International Journal of Hydrogen Energy. 37 (20): 15070–15080. doi:10.1016/j.ijhydene.2012.07.130. ISSN 0360-3199.
- Cha MS, Kim KH, Jo SJ, Lee NE, Lee JE, Lee JD, Lee SJ, Park JR (December 2003). "Identification and Characteristics of a Purple, Non-Sulfur Bacterium, Rhodobacter sp. EGH-24 from Korea Coast". Journal of the Environmental Sciences (in Korean). 12 (12): 1293–1301. doi:10.5322/jes.2003.12.12.1293. ISSN 1225-4517.
- Srinivas TN, Kumar PA, Sasikala C, Ramana CV, Süling J, Imhoff JF (July 2006). "Rhodovulum marinum sp. nov., a novel phototrophic purple non-sulfur alphaproteobacterium from marine tides of Visakhapatnam, India". International Journal of Systematic and Evolutionary Microbiology. 56 (Pt 7): 1651–1656. doi:10.1099/ijs.0.64005-0. PMID 16825644.
- Hiraishi, Akira; Nagao, Nobuyoshi; Yonekawa, Chinatsu; Umekage, So; Kikuchi, Yo; Eki, Toshihiko; Hirose, Yuu (2020-01-22). "Distribution of Phototrophic Purple Nonsulfur Bacteria in Massive Blooms in Coastal and Wastewater Ditch Environments". Microorganisms. 8 (2): 150. doi:10.3390/microorganisms8020150. ISSN 2076-2607. PMC 7074854. PMID 31979033.
- Azad SA, Vikineswary S, Chong VC, Ramachandran KB (January 2004). "Rhodovulum sulfidophilum in the treatment and utilization of sardine processing wastewater". Letters in Applied Microbiology. 38 (1): 13–18. doi:10.1046/j.1472-765x.2003.01435.x. PMID 14687209. S2CID 29239661.
- Azbar N, Levin DB (2011). "Biohydrogen production from Agricultural Agrofood-based resources". Comprehensive Biotechnology: 532–544. doi:10.1016/b978-0-444-64046-8.00381-5. ISBN 978-0-444-64047-5.
- Heinze C, Meyer S, Goris N, Anderson L, Steinfeldt R, Chang N, et al. (2015-06-09). "The ocean carbon sink – impacts, vulnerabilities and challenges". Earth System Dynamics. 6 (1): 327–358. doi:10.5194/esd-6-327-2015. hdl:1956/12384. ISSN 2190-4987. S2CID 17455839.
- Van Dusen H (November 2022). Kim C (ed.). "Using Microorganisms to Fight a Macro-level Problem". Princeton Public Health Review. Retrieved 2023-04-05.
- Engelhardt T, Sahlberg M, Cypionka H, Engelen B (August 2011). "Induction of prophages from deep-subseafloor bacteria". Environmental Microbiology Reports. 3 (4): 459–465. doi:10.1111/j.1758-2229.2010.00232.x. PMID 23761308.
- Heinrichs ME, Heyerhoff B, Arslan-Gatz BS, Seidel M, Niggemann J, Engelen B (2022-05-27). "Deciphering the Virus Signal Within the Marine Dissolved Organic Matter Pool". Frontiers in Microbiology. 13: 863686. doi:10.3389/fmicb.2022.863686. PMC 9184803. PMID 35694303.
- Talukdar C, Sastri S (2021-03-31). "Super Bacteria: A New Hope of Manufacturing Spider Silk in an Efficient Way". International Journal for Research in Applied Sciences and Biotechnology. 8 (2): 225–226. doi:10.31033/ijrasb.8.2.28. ISSN 2349-8889. S2CID 234840836.
- Acquavia MA, Pascale R, Martelli G, Bondoni M, Bianco G (January 2021). "Natural Polymeric Materials: A Solution to Plastic Pollution from the Agro-Food Sector". Polymers. 13 (1): 158. doi:10.3390/polym13010158. PMC 7796273. PMID 33406618.
- Carlozzi P, Touloupakis E (May 2021). "Bioplastic production by feeding the marine Rhodovulum sulfidophilum DSM-1374 with four different carbon sources under batch, fed-batch and semi-continuous growth regimes". New Biotechnology. 62: 10–17. doi:10.1016/j.nbt.2020.12.002. PMID 33333263. S2CID 229317407.
- Carlozzi P, Giovannelli A, Traversi ML, Touloupakis E (2021-02-01). "Poly(3-hydroxybutyrate) bioproduction in a two-step sequential process using wastewater". Journal of Water Process Engineering. 39: 101700. doi:10.1016/j.jwpe.2020.101700. ISSN 2214-7144. S2CID 226354079.
- Amstutz V, Hanik N, Pott J, Utsunomia C, Zinn M (2019). "Tailored biosynthesis of polyhydroxyalkanoates in chemostat cultures". In Bruns N, Loos K (eds.). Enzymatic Polymerizations. Methods in Enzymology. Vol. 627. pp. 99–123. doi:10.1016/bs.mie.2019.08.018. ISBN 9780128170953. PMID 31630749. S2CID 204813406.
- Higuchi-Takeuchi M, Motoda Y, Kigawa T, Numata K (August 2017). "Class I Polyhydroxyalkanoate Synthase from the Purple Photosynthetic Bacterium Rhodovulum sulfidophilum Predominantly Exists as a Functional Dimer in the Absence of a Substrate". ACS Omega. 2 (8): 5071–5078. doi:10.1021/acsomega.7b00667. PMC 6044645. PMID 30023736.
- Maeda, Isamu; Miyasaka, Hitoshi; Umeda, Fusako; Kawase, Masaya; Yagi, Kiyohito (2003-02-20). "Maximization of hydrogen production ability in high-density suspension ofRhodovulum sulfidophilum cells using intracellular poly(3-hydroxybutyrate) as sole substrate". Biotechnology and Bioengineering. 81 (4): 474–481. doi:10.1002/bit.10494. ISSN 0006-3592. PMID 12491532.
- Keefe AD, Pai S, Ellington A (July 2010). "Aptamers as therapeutics". Nature Reviews. Drug Discovery. 9 (7): 537–550. doi:10.1038/nrd3141. PMC 7097324. PMID 20592747.
- Hwang SD, Midorikawa N, Punnarak P, Kikuchi Y, Kondo H, Hirono I, Aoki T (December 2012). "Inhibition of Hirame rhabdovirus growth by RNA aptamers". Journal of Fish Diseases. 35 (12): 927–934. doi:10.1111/jfd.12000. PMID 22943666.
- Chong C, Low C (March 2019). "Synthetic antibody: Prospects in aquaculture biosecurity" (PDF). Fish & Shellfish Immunology. 86: 361–367. doi:10.1016/j.fsi.2018.11.060. PMID 30502461. S2CID 56479766.
- Balcázar JL, de Blas I, Ruiz-Zarzuela I, Cunningham D, Vendrell D, Múzquiz JL (May 2006). "The role of probiotics in aquaculture". Veterinary Microbiology. 114 (3–4): 173–186. doi:10.1016/j.vetmic.2006.01.009. PMID 16490324.
- Chang BV, Liao CS, Chang YT, Chao WL, Yeh SL, Kuo DL, Yang CW (2019-03-28). "Investigation of a Farm-scale Multitrophic Recirculating Aquaculture System with the Addition of Rhodovulum sulfidophilum for Milkfish (Chanos chanos) Coastal Aquaculture". Sustainability. 11 (7): 1880. doi:10.3390/su11071880. ISSN 2071-1050.
Further reading
- de Bont JA, Scholten A, Hansen TA (January 1981). "DNA-DNA hybridization of Rhodopseudomonas capsulata, Rhodopseudomonas sphaeroides and Rhodopseudomonas sulfidophila strains". Archives of Microbiology. 128 (3): 271–274. doi:10.1007/BF00422528. PMID 6971081. S2CID 19309759.
- Ormerod JG (1983). The Phototrophic bacteria: anaerobic life in the light. Berkeley: University of California Press. ISBN 978-0-520-05092-1.
- Falkow S, Dworkin M (2006). The prokaryotes: a handbook on the biology of bacteria. Berlin: Springer. ISBN 978-0-387-25495-1.