REEP5
Receptor expression-enhancing protein 5 is a protein that in humans is encoded by the REEP5 gene.[5][6][7] Receptor Expression Enhancing Protein is a protein encoded for in Humans by the REEP5 gene.
| REEP5 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | REEP5, C5orf18, D5S346, DP1, TB2, YOP1, Yip2e, receptor accessory protein 5, POB16 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 125265 MGI: 1270152 HomoloGene: 68479 GeneCards: REEP5 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
REEP5 is located on chromosome 5 between base pairs 112876385 to 112922289 on the minus strand.[8] The gene includes five exons.[8] The genes DCP2 and SRP19 are located upstream and downstream of REEP5 in humans.[9]
Protein
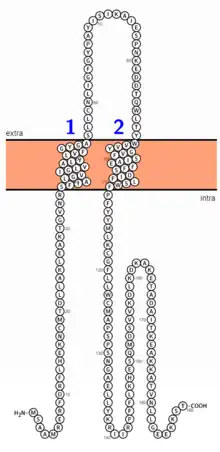
The protein is a member of the REEP family, which generally facilitate intracellular trafficking through alterations to the endoplasmic reticulum, and which have the ability to enhance activity of G-protein coupled receptors.[10] The human protein is 189 amino acids in length, containing two transmembrane regions and one named region- TB2_DP1_HVA22,[11] The pre-modification protein mass is 21.5 kdal.[12] Compared to the SwissProt collection of human proteins, REEP5 is composed of normal percentages of all amino acids.[12] Aside from a long stretch of electrically neutral amino acids, no significant patterns appear.[12] A second isoform exists in humans, and is 131 amino acids in length as the result of a change after the 117th amino acid.[13] Most of the fifth exon is not included.
Regulation
Tissue expression
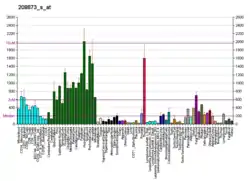
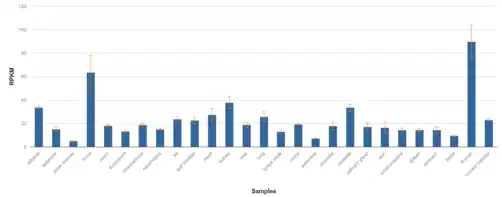
REEP5 is expressed across a number of tissues at a relatively high level, with at least 20 Reads Per Kilobase of transcript, per Million mapped reads appearing in adrenal, fat, gall bladder, heart, kidney, prostate, lung, and urinary bladder tissues.[14] Expression is even more highly elevated in brain and thyroid tissues.[14]
Transcription factors
EGR/Nerve Growth factor is a transcription factor with a potential binding site in the REEP5 promoter, which promotes neural plasticity, and therefore potentially provides an explanation for elevated levels of Receptor Expression Enhancing Protein 5 in the brain.[15]
Additionally, two transcription factors related to heart development and function have possible binding sites in the promoter. Transcription Box 20 (TBX20) is related to heart development, with protein deficiencies linked to poor heart function in adult mice, and death in infants before gestation.[16] MyoD is a Myogenic regulator which has been detected in the embryonic hearts of some birds in Purkinje cells, which are related to contractile function.
Protein level
At the sub cellular level, REEP5 is expressed in the endoplasmic reticulum.[17] Immunochemical staining localizes it here.[18]
A number of post-translational modifications are computationally predicted in humans and close orthologs. Acetylation of the second amino acid was predicted.[19] Phosphorylation of the 150th amino acid is predicted in Humans, Mice and Chickens.[20] O-GlcNac additions are also predicted at sites 128, 188, and 189.[21] Sumoylation sites are predicted at amino acids 147, 175, 186, and 187.[22] Propeptide cleavage was detected at amino acid 11.[23] Glycation was predicted at amino acids 16, 117, 164, 186, and 187.[24]
Homology
Paralogs
REEP5 has five paralogs in humans; REEP1, REEP2, REEP3, REEP4, and REEP[25]
| Paralog Name | Accession Number | Sequence Identity | Expected Millions of Year since divergence |
|---|---|---|---|
| REEP6 | NP_612402.1 | 54% | 563.16 |
| REEP4 | AAH50622 | 25% | 1325.64 |
| REEP1 | NP_001303894.1 | 24.6% | 1341.62 |
| REEP3 | AAT70686.1 | 22.7% | 1421.20 |
| REEP2 | NP_057690.2 | 15.6% | 1792.58 |
Orthologs
REEP5 has orthologs as far removed from humans as Symbiodinium microadriacticum, a protist species.[25] Sequence identity is high for vertebrates, but decreases significantly outside of this group. The DP1/TB2/HVA22 region is well conserved compared to the two transmembrane regions.
| Organism | Date of Divergence (millions of years ago) | Taxonomic Group | Sequence Accession Number | Sequence identity | e-value |
|---|---|---|---|---|---|
| Homo Sapiens | NA | Mammalia | NP_005660.4 | 100% | NA |
| Mus musculus | 89 | Mammalia | N_031900.3 | 93.7% | 4x10−134 |
| Danio rerio | 433 | Actinopterygii | NP_956352.1 | 72.0% | 7x10−104 |
| Drosophilia melanaogaster | 736 | Insecta | NP_001188928.1 | 38.2% | 9x10−57 |
| Jaminia rosea | 1017 | Exbasidiomycetes | XP_025360265 | 31.9% | 5x10−40 |
| Rhodotorula graminis WP1 | 1105 | Microbotryomycetes | XP_018269688.1 | 31.2% | 7x10−35 |
| Citrus unshiu | 1275 | Magnoliopsida | Gay51937.1 | 25.2% | 4x10−26 |
| Symbiodinium microadriacticum | 1552 | Dinophyceae | OLQ12882.1 | 16.2% | 5x10−16 |
Evolutionary Rate
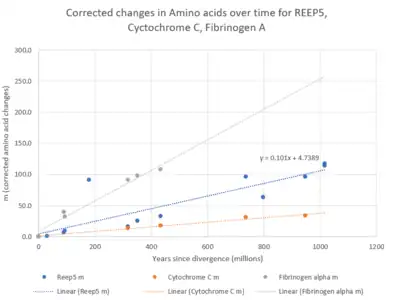
The evolutionary rate of the REEP5 protein is moderately high, falling between that of cytochrome c and fibrinogen alpha. Neutral mutations are expected to occur around once per one million years
Interacting partners
REEP5 interacts with a number of proteins, including Regulator of G Protein signalling 2, which was detected using Yeast 2 Hybridization. RGS2 plays a role in G protein signalling 2 pathways and the contraction of smooth muscle tissues. Derlin 2 (Derl2) interacts with REEP5 as detected by anti-tag coimmunoprecipation and degrades misfolded glycoproteins in the endoplasmic reticulum.[26] Cholinergic Receptor Muscarinic 5 was detected as an interactant by ubiquitin reconstruction.[27] CHRM5 binds acetylcholine, and can impact the function of the central and peripheral nervous systems.[28]
Regulator of G Protein Signaling 2 was also detected with Yeast 2 Hybridization and is a regulator of G-protein receptor which may be involved with the contraction of smooth vascular muscle.[27][29]
Four proteins were experimentally detected across several papers.[30] Three of them, Atlastin GTPase 1, Atlastin GTPase 2, and Zinc Finger FYVE-Type Containing 27 are related to the structuring of the endoplasmic reticulum (tubules in particular), the development of axons, or neuron growth.[17][31] The fourth is REEP6, which is responsible for transporting receptors to the cell surface, as well as regulating endoplasmic reticulum structure.[32]
SARS-CoV-2
REEP5 is one of many human proteins which interacts with SARS-CoV-2. Specifically, the coronavirus protein P0DTC5, which appears in the intermediate compartment between the endoplasmic reticulum and golgi apparatus, interacts with REEP5. P0DTC5 is a viral envelope protein which is crucial to viral morphogenesis, making it possible that REEP5 is related in someway to packaging or release of the virus from human cells.[33]
Clinical significance
Research has linked REEP5 to several diseases and disorders.
In colon cancer, expression of REEP5 enhances activity of CXCR1, a protein receptor which contributes to the growth and spread of cancer cells. Researchers have found that under expression of REEP5 reduces the efficiency of CXRC1, and reduces the ability of lung cancer cells to metastasize.[10] This provides both a potential target for cancer therapy and further evidence of REEP5 having function as a protein which increases activity of other receptor proteins. Another type of cancer related to REEP5 is colon cancer, in which the protein normally interacts with and neutralizes HCCR1, a protein which would otherwise interfere with P53 and contribute to the progression of cancer.[34]
REEP5 has also been linked to heart disorders, and research into the connection has revealed that it helps organize the junctional Sarcoplasmic Reticulum in myocytes. REEP5 aligns the junctional SR to T-tubules, which promotes effective release of Calcium ions. When protein function is disrupted the junction becomes misaligned and Calcium ion release becomes less effective. Given the importance of Calcium ions to the muscle contractions which power the heart, targeting REEP5 has the potential to be a therapeutic option in some disorders.[35]
Additionally, there is some statistical evidence linking certain SNPs found within REEP5 mRNA to Major Depressive Disorder.[36] However, a biological mechanism has not been confirmed, and the study only looked at a limited population.[36]
References
- GRCh38: Ensembl release 89: ENSG00000129625 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000005873 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Clark AJ, Metherell LA, Cheetham ME, Huebner A (December 2005). "Inherited ACTH insensitivity illuminates the mechanisms of ACTH action". Trends in Endocrinology and Metabolism. 16 (10): 451–7. doi:10.1016/j.tem.2005.10.006. PMID 16271481. S2CID 27450434.
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H (November 2004). "RTP family members induce functional expression of mammalian odorant receptors". Cell. 119 (5): 679–91. doi:10.1016/j.cell.2004.11.021. PMID 15550249. S2CID 13555927.
- "Entrez Gene: REEP5 receptor accessory protein 5".
- "REEP5 receptor accessory protein 5 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-07-30.
- "Human hg38 chr5:112,865,773-113,003,301 UCSC Genome Browser v401". genome.ucsc.edu. Retrieved 2020-07-31.
- Park CR, You DJ, Park S, Mander S, Jang DE, Yeom SC, et al. (December 2016). "The accessory proteins REEP5 and REEP6 refine CXCR1-mediated cellular responses and lung cancer progression". Scientific Reports. 6: 39041. Bibcode:2016NatSR...639041P. doi:10.1038/srep39041. PMC 5155276. PMID 27966653.
- "receptor expression-enhancing protein 5 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-06-22.
- "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2020-07-31.
- "UniProtKB - Q00765 (REEP5_HUMAN)". UniProt. 2020-06-17. Retrieved 2020-07-30.
- "REEP5 Gene Expression - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-07-30.
- Levkovitz Y, Baraban JM (May 2002). "A dominant negative Egr inhibitor blocks nerve growth factor-induced neurite outgrowth by suppressing c-Jun activation: role of an Egr/c-Jun complex". The Journal of Neuroscience. 22 (10): 3845–54. doi:10.1523/JNEUROSCI.22-10-03845.2002. PMC 6757642. PMID 12019303.
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, et al. (May 2005). "Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation". Development. 132 (10): 2451–62. doi:10.1242/dev.01799. PMID 15843414.
- Chang J, Lee S, Blackstone C (September 2013). "Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation". Proceedings of the National Academy of Sciences of the United States of America. 110 (37): 14954–9. Bibcode:2013PNAS..11014954C. doi:10.1073/pnas.1307391110. PMC 3773775. PMID 23969831.
- "REEP5] Primary Antibodies". www.thermofisher.com. Retrieved 2020-07-31.
- "NetAcet 1.0 Server". www.cbs.dtu.dk. Retrieved 2020-08-01.
- "NetPhos 3.1 Server". www.cbs.dtu.dk. Retrieved 2020-08-01.
- "YinOYang 1.2 Server". www.cbs.dtu.dk. Retrieved 2020-08-01.
- "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". sumosp.biocuckoo.org. Retrieved 2020-08-01.
- "ProP 1.0 Server". www.cbs.dtu.dk. Retrieved 2020-08-01.
- "NetGlycate 1.0 Server". www.cbs.dtu.dk. Retrieved 2020-08-01.
- "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2020-07-31.
- Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K (January 2006). "Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation". The Journal of Cell Biology. 172 (3): 383–93. doi:10.1083/jcb.200507057. PMC 2063648. PMID 16449189.
- "The Molecular INTeraction Database – An ELIXIR Core Resource". Retrieved 2020-08-02.
- "CHRM5 cholinergic receptor muscarinic 5 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-08-02.
- "RGS2 regulator of G protein signaling 2 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-08-02.
- "PSICQUIC View". www.ebi.ac.uk. Retrieved 2020-08-02.
- "ZFYVE27 zinc finger FYVE-type containing 27 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2020-08-02.
- Reference, Genetics Home. "REEP6 gene". Genetics Home Reference. Retrieved 2020-08-02.
- "InterPro". www.ebi.ac.uk. Retrieved 2020-08-02.
- Shin SM, Chung YJ, Oh ST, Jeon HM, Hwang LJ, Namkoong H, et al. (June 2006). "HCCR-1-interacting molecule "deleted in polyposis 1" plays a tumor-suppressor role in colon carcinogenesis". Gastroenterology. 130 (7): 2074–86. doi:10.1053/j.gastro.2006.03.055. PMID 16762630.
- Yao L, Xie D, Geng L, Shi D, Huang J, Wu Y, et al. (February 2018). "REEP5 (Receptor Accessory Protein 5) Acts as a Sarcoplasmic Reticulum Membrane Sculptor to Modulate Cardiac Function". Journal of the American Heart Association. 7 (3): e007205. doi:10.1161/JAHA.117.007205. PMC 5850239. PMID 29431104.
- Yang Z, Ma X, Wang Y, Wang J, Xiang B, Wu J, et al. (2012-09-01). "Association of APC and REEP5 gene polymorphisms with major depression disorder and treatment response to antidepressants in a Han Chinese population". General Hospital Psychiatry. 34 (5): 571–7. doi:10.1016/j.genhosppsych.2012.05.015. PMID 22795047.
Further reading
- Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. (August 1991). "Identification of FAP locus genes from chromosome 5q21". Science. 253 (5020): 661–5. Bibcode:1991Sci...253..661K. doi:10.1126/science.1651562. PMID 1651562.
- Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, et al. (August 1991). "Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients". Science. 253 (5020): 665–9. Bibcode:1991Sci...253..665N. doi:10.1126/science.1651563. PMID 1651563.
- Spirio L, Joslyn G, Nelson L, Leppert M, White R (November 1991). "A CA repeat 30-70 KB downstream from the adenomatous polyposis coli (APC) gene". Nucleic Acids Research. 19 (22): 6348. doi:10.1093/nar/19.22.6348. PMC 329171. PMID 1659692.
- Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, Samowitz W, et al. (August 1991). "Identification of deletion mutations and three new genes at the familial polyposis locus". Cell. 66 (3): 601–13. doi:10.1016/0092-8674(81)90022-2. PMID 1678319. S2CID 33965913.
- Prieschl EE, Pendl GG, Harrer NE, Baumruker T (March 1996). "The murine homolog of TB2/DP1, a gene of the familial adenomatous polyposis (FAP) locus". Gene. 169 (2): 215–8. doi:10.1016/0378-1119(95)00827-6. PMID 8647449.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. (January 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Shin SM, Chung YJ, Oh ST, Jeon HM, Hwang LJ, Namkoong H, et al. (June 2006). "HCCR-1-interacting molecule "deleted in polyposis 1" plays a tumor-suppressor role in colon carcinogenesis". Gastroenterology. 130 (7): 2074–86. doi:10.1053/j.gastro.2006.03.055. PMID 16762630.